Main Text
1 Introduction
Thrombosis, the blocking of blood vessels by blood clots, along with the related thrombo-inflammation and thromboembolism, is a chief cause of cardiovascular disease [1-6]. Consequently anything that can promote safe anti-coagulation or fibrinolytic activity is likely to have therapeutic potential (e.g., [7-15]).
Nattō (usually rendered natto) is a Japanese food made via the fermentation of soy beans using the Gram-positive organism Bacillus subtilis var natto [16-20]. It has been widely consumed for over 2000 years, and is considered safe [21]. The proteolytic activity of natto was detected in 1906 [22] and its fibrinolytic activity in 1925 [23]. However, it was not until 1987 [21] that an enzyme exhibiting these activities was purified from natto; in spite of it being a protease it was termed nattokinase [21].
Despite having to pass through the gut wall [24-31], nattokinase is orally available (and this can be improved [32-34]), is considered a major contributor to the purported health benefits of natto [21,27,35-60], not least in cardiovascular disease [27,28,36,38,39,41,53,61-80], and is itself recognised as safe [64,81,82].
The experimental 3D structure of nattokinase, which is a serine protease related to subtilisin, is available [83,84], and it may also be produced via purification [85-88] or (as here) recombinantly [44,67,68,72,89-100]. Although not our prime focus in this paper, it is also known to cleave plasminogen activator inhibitor I [101], to have antiplatelet [102], anti-inflammatory [77], and anti-hypertensive [65,103,104] activities, and to show neuroprotective [105] and post-stroke benefits [106] as well, when dosed adequately, as having anti-lipidaemic effects [69].
Following earlier work using electron microscopy (e.g., [107-110]), we discovered that fibrinogen could polymerise or clot into an anomalous, amyloid form of fibrin (e.g., [111-118]) that exactly reflected the clots seen in both the electron microscope [119] and in bright field optical microscopy [120]. As with prions and other amyloid forms of proteins [112,121], that are often highly resistant to proteolysis (e.g., [122,123]), the existence of these ‘fibrinaloid’ microclots implies their comparative resistance to normal fibrinolysis [124,125], with their precise structures [126] being affected by other small and macromolecules and ions that they may have bound [111,117,127-133]. The varieties of stable macrostates into which a given amyloidogenic sequence can fold (even under the same conditions [134,135]) are referred to as different ‘strains’ [136-146] or ‘polymorphisms’ [147-158], and in some cases are sufficiently stable (i.e., kinetically isolated from other macrostates) that they are even heritable [136,159-165]. Homo- and hetero-polymerisation and their catalysis are then referred to, respectively, as (self-)‘seeding’ [154,166-180] and ‘cross-seeding’ [167,181-188]. More recently, we have established the prevalence of these fibrinaloid microclots in post-viral diseases such as Long COVID [120,189-192] (and see confirmation by others in [193,194]) and ME/CFS (myalgic encephalopathy/chronic fatigue syndrome) [195,196]. The lower amyloidogenicity of omicron versus earlier variants of SARS-CoV-2 is also reflected in its lower virulence [197], implying that these microclots are on the aetiological pathway of the disease, and they can explain many symptoms [198], including fatigue [199], post-exertional symptom exacerbation [200], autoantibody generation [121] and Postural Orthostatic Tachycardia Syndrome (POTS) [201]. Fibrin amyloid microclots also occur during sepsis [202], while amyloid deposits are also observed in the skeletal muscles of those with Long COVID [203]. Overall, this ability of fibrinaloid microclots to provide a mechanistic explanation of multiple phenomena is consistent with the ‘explanatory coherence’ view of science [204-207]. In common with other amyloid proteins [112], that contain a characteristic cross-β motif [187,208-224], they can be visualized using the fluorogenic stain thioflavin T [158,187,225-239] or via vibrational spectroscopy [236,240-251]. As with any other ligand or binding agent, the rotation of the bound form is more restricted than that of the free form (which is largely what makes it fluorogenic), and precise intensities of thioflavin T fluorescence depend on the location and conformation(s) to which the thioflavin T is bound [227-229,252-269] and in some cases on the presence of interferents [270].
Although nattokinase preparations are widely available commercially, and as noted above they are considered to have significant therapeutic value, including in Long COVID [271,272], their exact contents are uncertain, and so we decided that it was best to create and use purified, recombinant material.
While the proteolytic specificity of nattokinase, as an alkaline serine protease [44,73,273], is surprisingly underexplored, beyond a broad similarity to that of plasmin [44,274] (and nattokinase can even degrade spike protein [275] and certain ‘classical’ amyloids [276-278]), the question arises as to whether or not nattokinase can degrade the amyloid ‘fibrinaloid’ form of microclots. The purposes of this paper are (i) to describe an efficient, quantitative, automated microscopic method that can be used to determine the size and number of amyloid microclots and any time-dependent changes therein, and thus (ii) to assess any such nattokinase-induced degradation of the microclots, concluding that nattokinase can indeed degrade fibrinaloid microclots effectively. The therapeutic implications of this are discussed.
2 Materials and methods
2.1 Assay method
In vitro microclots were made by mixing 45 μL commercially obtained fibrinogen (Sigma catalogue number 9001-32-5, at a final concentration of 2 mg/mL) with 5 μL bacterial LPS (Sigma product code L2630-10MG) and used at a final concentration of 1 ng/mL were incubated at 37 ºC for 15 min. 25 μL were removed and replaced with 25 μL thrombin (Sigma, final amount 14U) and incubated at 37 ºC for a further 15 min. 3 μL were removed and replaced with 3 μL of the fluorogenic amyloid dye, Thioflavin T (ThT) (final concentration: 0.03 mM) and incubated for 20 min (protected from light) at room temperature. Following incubation, 10 μL of the recombinantly produced nattokinase at different concentrations / PBS (control) were added. This was then followed by immediately pipetting 15 μL of assay sample onto a 15-well slide ‘angiogenesis’ glass bottom plate used without a lid (Ibidi: https://ibidi.com/chambered-coverslips/245--slide-15-well-3d-glass-bottom.html), reproduced in Figure 1, and without shaking (cf. [156,279-291]). The excitation wavelength band for ThT was set at 450 nm to 499 nm and the emission at 499 nm to 529 nm. Samples were viewed using Gen5 software on an Agilent BioTek Cytation 1 Cell Imaging Multimode Reader, essentially following the protocol developed and described by Dalton and colleagues [193]. The Cytation instrument is an automated fluorescence microscope with 8-bit intensity resolution in which an entire, large field of view can be constructed at high magnification by taking serial images and moving the stage automatically. With the 4× objective used, each final image (as in Figure 1) was composed of 1,296 individual images. The typical file size of a final, stitched .tif image was 19 Mb. Each experiment was run multiple times, each time being in triplicate (three separate wells). Other relevant settings that we optimized for this assay were as follows: the Cytation 1 temperature was set at 37 ºC, and images were taken every 41 min for 6 hours. The colour channel used was GFP 469,525. A fixed focal height setting, with a bottom elevation of 549 μm and 0 μm offset was selected. A Z-Stack montage of the entire well was applied, with a step size of 86.9 μm, and 12 slices. Samples were analysed using the Gen5 Image Prime 3.13.15 software supplied with the instrument, and the thresholds for minimum and maximum object (clots) size that could be detected were set at 5 and 500 μm, respectively.

Figure 1 lbidi 15-well 3D glass-bottomed microslide as used herein (the incubation system used herein, allowing imaging from below).
2.2 Recombinant nattokinase
Recombinant nattokinase was produced within the Liverpool Gene Mill. The nucleotide sequence for Bacillus subtilis nattokinase (Uniprot Q93L66, GenBank: AER52006.1) was synthesised by Twist Bioscience and supplied in the pET28a(+) plasmid. The sequence was modified to include a C-terminal poly-Histidine tag for purification, as well as an N-terminal PelB leader sequence in which the terminal QPAMA residues are replaced by APOIA, and with a penta-aspartate linker for targeting to the periplasmic space [292] plus a ENLYFQ TEV cleavage site and a further SGS linker prior to the nattokinase sequence (beginning AQSVPY). The vector was used to transform chemically competent cells of the RosettaTM strain of Escherichia coli (Novagen) according to the method described by Inoue et al. [293]. Transformed cells were plated on plates of LB-agar (0.5% w/v yeast extract, 1% w/v NaCl, 1% w/v tryptone and 2% agar) supplemented with 50 μg/mL kanamycin and 25 μg/mL chloramphenicol. A single colony from the agar plate was used to inoculate 5 mL of LB broth (0.5% w/v yeast extract, 1% w/v NaCl, 1% w/v tryptone) supplemented with kanamycin and chloramphenicol as described above, for overnight culturing at 37 °C with shaking. The culture was diluted to an OD600 of 0.05 in 500 mL of LB broth supplemented with kanamycin and chloramphenicol as described above, and incubated with shaking at 37 °C. When an OD600 of 0.6 was reached, recombinant protein expression was induced by addition of 0.75 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and the culture was incubated overnight at 18 °C with shaking. Cell pellets were harvested by centrifugation at 4,000× for 10 min, and the pellets were resuspended in 50 mL of a solution of Tris-HCl (pH8) and 10 mM EDTA, and incubated at 60 °C for 2 hours [294]. The suspension was centrifuged at 4 °C at 16,000× for 10 min, and the supernatant was passed through 1 mL of HisPurTM Ni-NTA resin (Thermo Scientific) to purify poly-Histidine-tagged proteins. Bound proteins were eluted using 500 mM imidazole, followed by desalting and concentration using a PierceTM Protein Concentrator PES (Thermo Scientific) with 30 kDa cut-off. Protein yield was quantified using the PierceTM Bradford Protein Assay Kit (Thermo Scientific), and samples were frozen with 10% v/v glycerol until further use. Inclusion body formation [295] was not here a significant issue. Figure 2 shows a gel illustrating the final preparation.

Figure 2 SDS-PAGE of recombinant Nattokinase. F: sample flow-through (unbound proteins); W: fraction of wash buffer (100 mM Tris, pH 7.5, 150 mM NaCl, 50 mM imidazole); 1-3: purifed fractionsusing elution buffer (100 mM Tris, pH 7.5, 150 mM NaCl, 500 mM imidazole); C1 & C2: purified samples concentrated through 30 kD cut-off protein concentrator unit; C3: C1 & C2 samples pooled and further concentrated through 3 kD cut-off protein concentrator unit.
A kinetic experiment was set up on the Cytation 1 and the effect of nattokinase on microclots was studied at final concentrations of 28 ng/μL and 14 ng/μL, using ThT at a final concentration of 0.005 mM, as the fluorogenic dye. Measurements were taken every 40 min.
3 Results
3.1 Basic phenomenon, and effect of concentration of NK and incubation time
To give an indication of the kinds of data obtained in this study, Figure 3 (left panels) shows three Cytation images representing clots as stained with thioflavin T following incubation of fibrinogen plus thrombin plus LPS (as in [111]) for 6 hours, either with no further additions (Figure 3A, top), with PBS (Figure 3B, middle), or after simultaneous exposure to 28 ng/μL nattokinase.
While we sought to avoid any ‘cherry picking’ in the past, the great attraction of the present approach is that the entire sample is imaged (serially) so this issue is completely avoided. Although not necessarily obvious to the naked eye, there are variations in pixel intensity that allow a thresholding to determine what counts as a clot boundary. Figure 3 also shows the pixel intensity variation for the images displayed on its left side; the logarithmic plot in particular makes clear how much the pixels of larger intensity differed following the addition of the nattokinase.

Figure 3 Images of fibrinaloid microclot formation and their removal via nattokinase. Thrombin and fibrinogen were incubated together with thioflavin T and LPS, and imaged after 6 hours in a Cytation 1, as described in Methods. Further additions were (A) none, (B) PBS, (C) recombinant nattokinase 28 ng/μL. Bar = 2 mm (2,000 μm).
The time evolution of these data (Figure 4) shows that in the absence of nattokinase the clot numbers increase for an hour or so then decrease slightly before stabilizing (Figure 4A). When nattokinase is present the clot numbers decrease after the first time point and by 2 hours have attained their lowest level, this being approximately half that of the 14 ng/μL nattokinase (in which the nattokinase level is thus halved), possibly implying a loss of activity over time. In Figure 4B we see the dynamics of the clot intensity (total number of pixels), this being substantially lower in the presence of NK, especially at the higher level of enzyme. Figure 4C shows the time evolution of the median clot size.

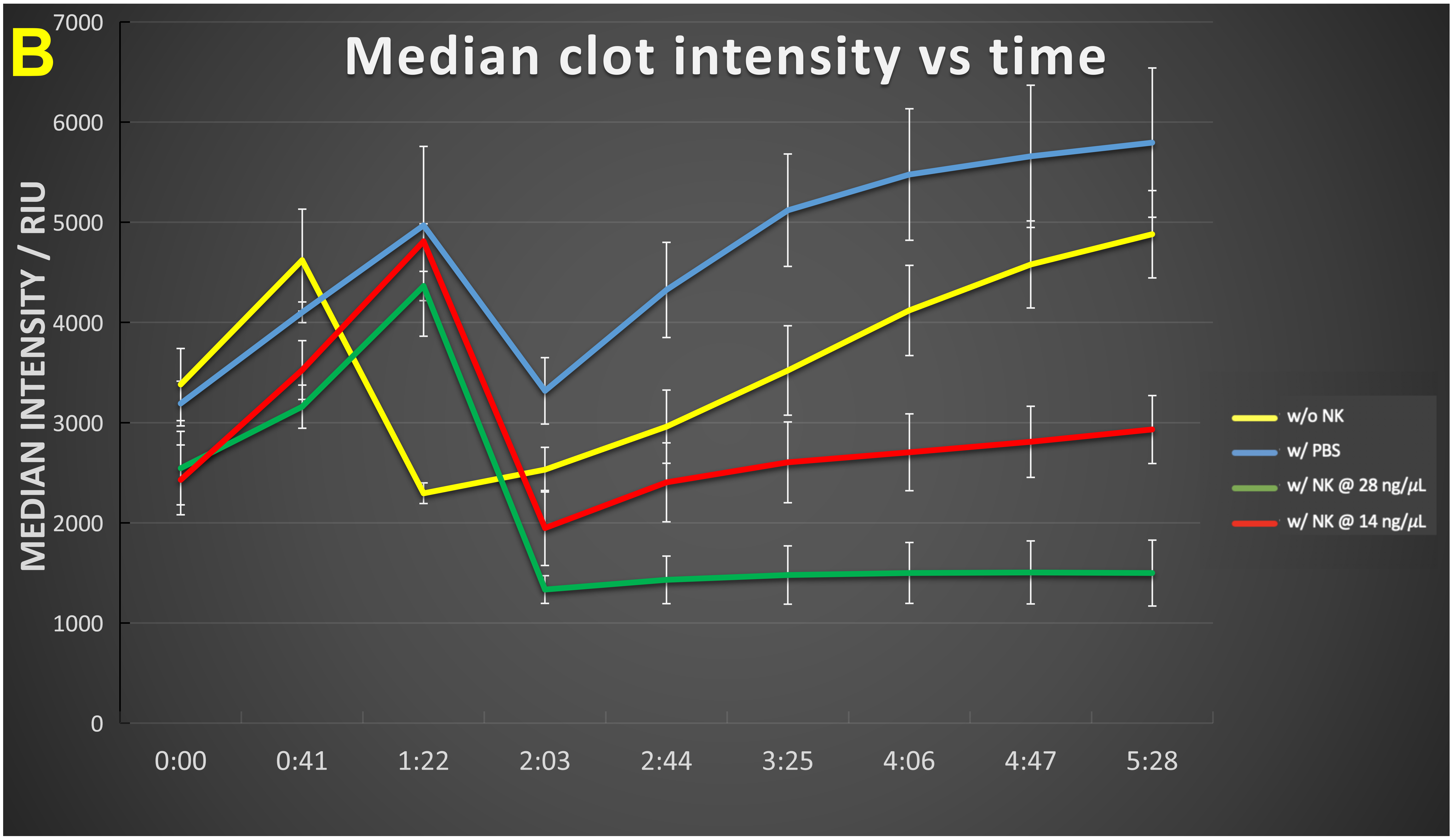
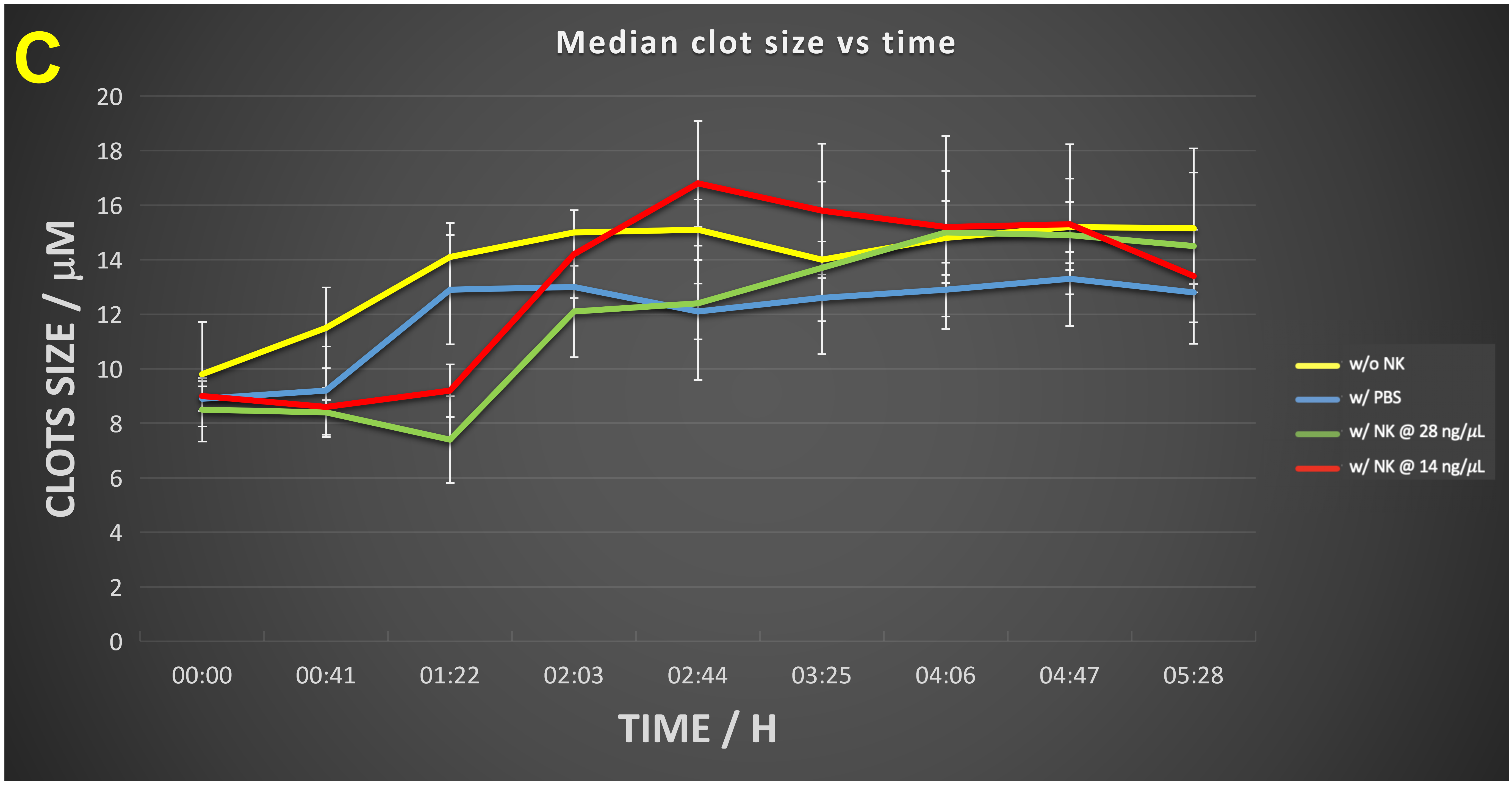
Figure 4 Time evolution of (A) clot number, (B) intensity and (C) median clot size during the development of fibrinaloid microclots and their incubation with nattokinase. Thrombin and fibrinogen were incubated together with thioflavin T, and imaged in a Cytation 1, as described in Methods. Further additions were none (yellow), PBS (blue), recombinant nattokinase 28 ng/μL (green), or recombinant nattokinase 14 ng/μL (red). Videos of the incubation with PBS and with nattokinase are given in Supplementary Information. Error bars represent standard deviations of triplicates within a single experiment (and the experiment was repeated on two separate days). The p-values (paired, one-tailed t-test) at 2 hours were as Table 1:
Table 1 p-values (paired, one-tailed t-test) at 2 hours.
| p-value | ||||
|---|---|---|---|---|
| w/o NK and w/ PBS | w/o NK and w/ NK at 28 ng/μL | w/o NK and w/ NK at 14 ng/μL | NK at 14 ng/μL vs. 28 ng/μL | |
| For median cell count | 0.35 | 0.0003 | 0.0014 | 0.055 |
| For median clot intensity | 0.004 | 0.0008 | 0.015 | 0.023 |
| For median clot size | 0.019 | 0.008 | 0.126 | 0.25 |
3.2 Effect of flowing conditions, as implemented by shaking
The above analyses were done online within the Cytation 1 throughout, and under static conditions. However, it is known that flow conditions—as would be the case in blood in vivo—can themselves stimulate amyloidogenic fibre formations [288-290,296-301]. It was thus of interest to compare (with individual time samples added to the Cytation 1), the effect of flow. To mimic this we used simple shaking (Figure 5). As indicated (Figure 5), shaking had a significant influence in decreasing the clot number (Figure 5A) while increasing the clot size (Figure 5B).
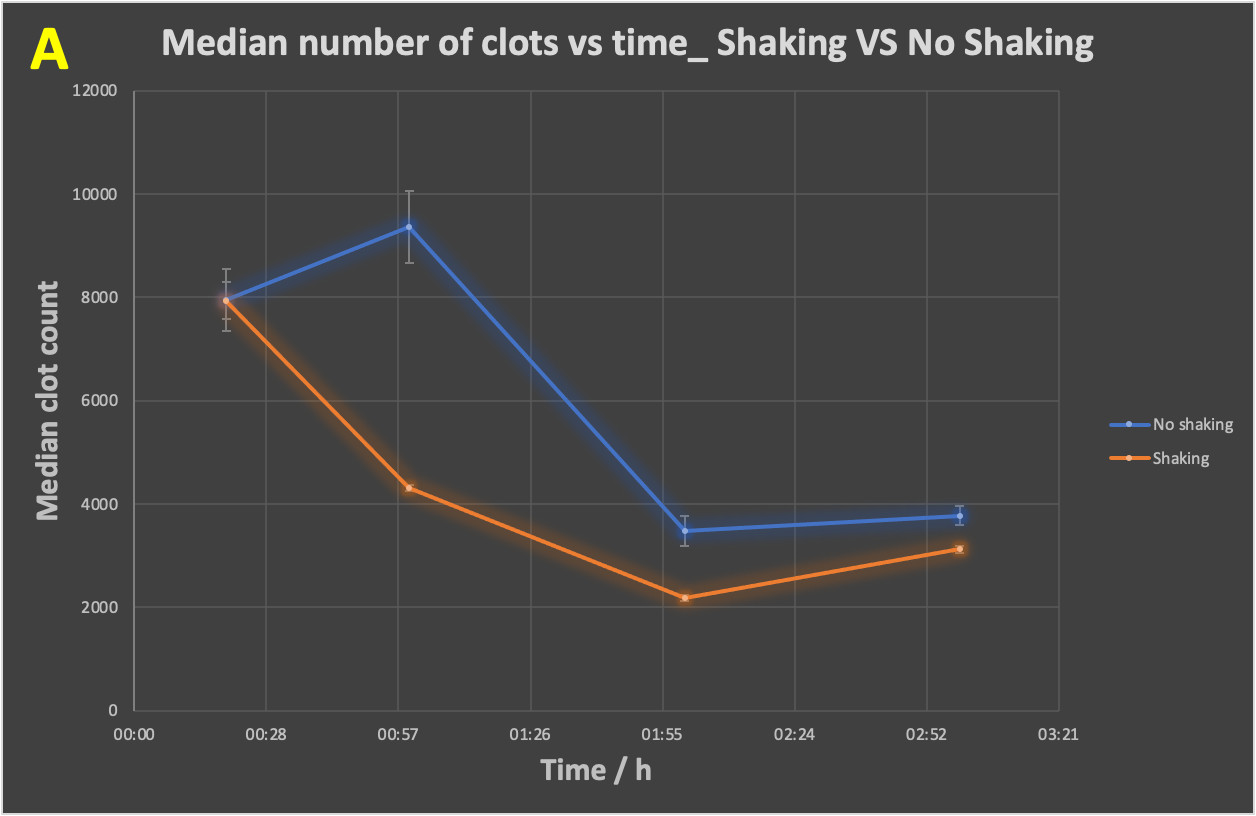
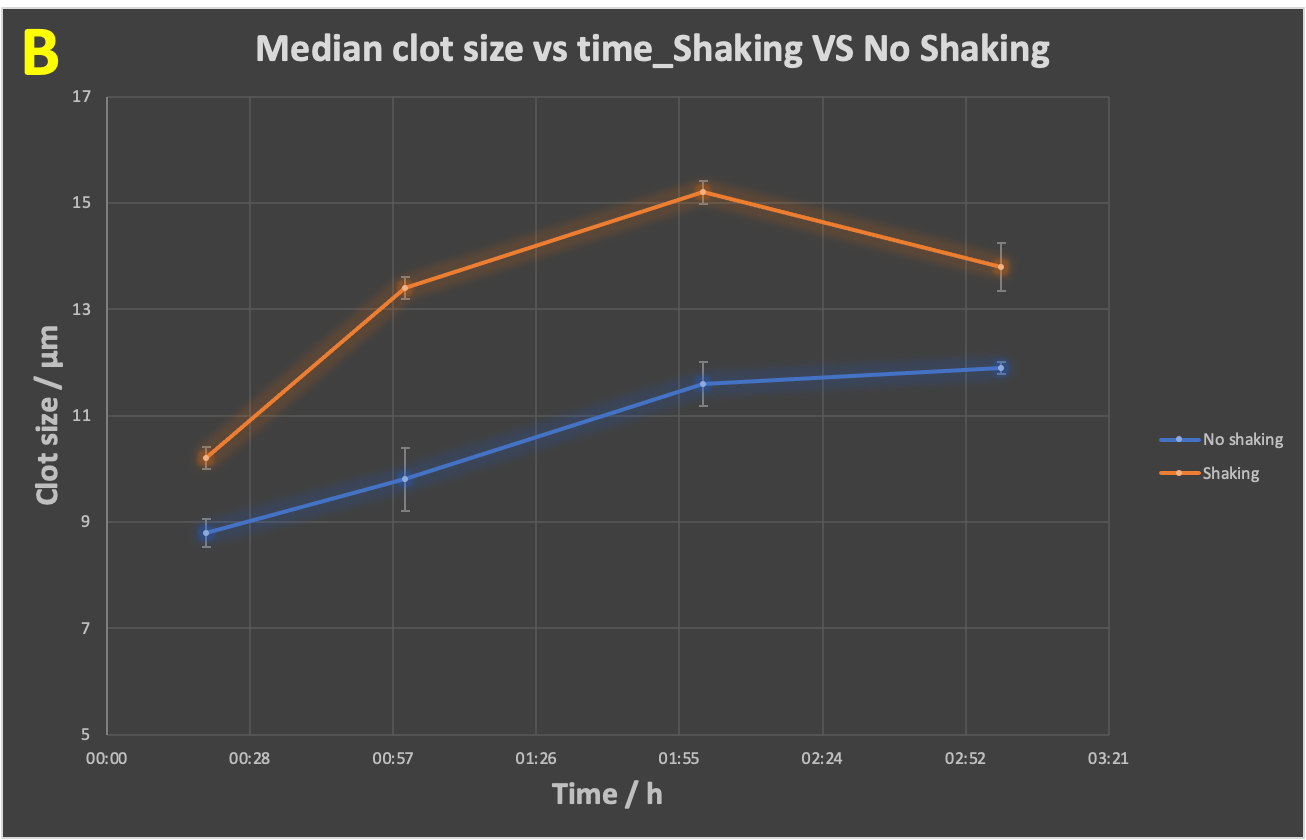
Figure 5 Time evolution of (A) clot number, (B) median clot size during the development of fibrinaloid microclots whilst shaking versus no shaking of the slides. Fibrinogen, thrombin and LPS were incubated together with thioflavin T, followed by immediate pipetting of 15 μL of assay sample onto a 15-well slide ‘angiogenesis’ glass bottom plate (as per Figure 1), which was left incubating for 20 mins, 1 hour, 2 hours and 3 hours, in a dark room, at 37 ºC, in a shaking incubator at 170 rpm. The no-shaking control samples were incubated under the same conditions but under stationary conditions. Slides were then imaged on the Cytation 1, using the GFP filter block, and the appropriate exposure settings. The p-value (paired, one-tailed t-test) for the difference in median clot size is 0.01. The experiment was duplicated, with each experiment having three technical replicates.
3.3 Using Amytracker dyes instead of ThT
Because it is valuable to have other dyes should one wish to use multiple wavelengths (as in [117]), we also assessed the red oligothiophene-class AmytrackerTM dyes (Ebba Biotech) (see e.g., [115,117,119,302-309]). However, these gave highly anomalous traces in this system, and given that they did previously stain the fibrinaloid microclots as mentioned in those references we suspect may have inhibited the nattokinase, so were not further pursued.
4 Discussion
Fibrinogen, especially its α-chain, is known to be amyloidogenic [112,310-316], and certain alleles are especially prone to causing fibrinogen-driven amyloidoses (e.g., [317-326]). Indeed, our own studies [111,113,115-118,120,191,197,199,327-333], and those of others [193,194,202], have demonstrated via staining with thioflavin T the authentic amyloidogenesis of fibrinogen to form fibrinaloid microclots. In the present work, we describe a medium throughput method for their quantitative estimation.
Three features stand out from the data in Figure 4. First, especially in the absence of NK (yellow trace), the clots increase in both number and size over time (Figure 4A,4B), illustrating how microclots may aggregate to form macroclots, as part of the normal amyloidogenic process (e.g., [187,227,230,254,312,334-344]), and such aggregation was increased under flow conditions (Figure 5). This kind of aggregation may be highly significant in stroke and myocardial infarctions, where clots may be far larger (e.g., [345-349]) than the simple sloughing off of atherosclerotic plaques might reasonably create. Secondly, the enzyme effectively decreases the rate and extent of microclot formation, in rough proportion to the amount of enzyme (compare e.g., red and green traces at 5 hours). The lowest intensity point was observed in the interval 2-4 hours, implying a die-off in activity or instability of the enzyme over time. This is good, in that untrammelled fibrinolytic activity may not be of the greatest therapeutic benefit. Lastly, the median clot size (Figure 4C) increases briefly then stabilizes. This reflects the fact that smaller clots will tend to be degraded preferentially as their surface area per unit mass is significantly greater than that of larger clots. (It is not commonly recognised, but if one imagines two solid spheres, of which one is twice the diameter of the other, the degradation of a given (i.e., the same) mass in the two spheres leads to a loss in mass of just 12.5% of the larger sphere when the smaller one is completely degraded, and a loss in radius of the larger sphere that is less than 5% of its starting value. Consequently, although possibly at first glance surprising, this is, given the traces in Figures 4A and 4B, in fact the result expected for Figure 4C.
The ability to assess the rate of fibrin amyloid formation and degradation noninvasively is highly desirable, as it precisely permits studies of the present type that can then be automated. While still not a high-throughput approach in the usual sense (flow clotometry [191,333], albeit using more expensive instrumentation, is certainly quicker), this does provide a substantial advance in scoring fibrinaloid microclot formation that is both fully quantitative and without undue operator fatigue. This has allowed us, for the first time, to conclude at least three important features: (i) the formation kinetics of fibrin amyloid microclots in whole samples may be imaged noninvasively in an automated manner, (ii) such microclots can aggregate over time, and (iii) the fibrinaloid microclots may be degraded by nattokinase. This latter has significant therapeutic implications for those suffering from Long COVID and related disorders, as NK preparations are widely available commercially. Our approach also thus allows for the comparison of different preparations of NK. Future work could usefully include recombinant serrapeptase (NK/SP), lumbrokinase (NK/LK) and/or sequence variants of NK/SP made using the methods of synthetic biology [350], since both serrapeptase and lumbrokinase, and even papain [351], also have fibrinolytic and amyloid-degrading properties [60,352-363].
Of course these results might also be recognized as having clinical potential. Given the evidence for the relevance of fibrinaloid microclots in the aetiology of Long COVID and other post-infection diseases as rehearsed above, and the potential shown here of such enzmes to remove them, a randomized controlled trial of these kinds of fibrinolytic enzymes (vs. placebo) seems more than warranted.
Back Matter
Acknowledgments
We thank Dr. Caroline F. Dalton (Sheffield Hallam University) and Drs. Amanda Barnes and Ashley Smith (Agilent) for many helpful discussions about the Cytation system and its optimisation.
Conflicts of Interest
E.P. is a named inventor on a patent application covering the use of fluorescence methods for microclot detection in Long COVID. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Author Contributions
Conceptualization, D.B.K., E.P.; methodology, J.M.G., J.E.S.-S., C.W.T.; resources, D.B.K., E.P.; writing—original draft preparation, D.B.K.; writing—review and editing, all authors; project administration, D.B.K., E.P.; funding acquisition, D.B.K., E.P. All authors have read and agreed to the published version of the manuscript.
Ethics Approval and Consent to Participate
The manuscript didn't involve any human or animal subjects, therefore no ethical approval was required for this article.
Funding
D.B.K. thanks the Balvi Foundation (grant 18) and the Novo Nordisk Foundation for funding (grant NNF20CC0035580). E.P.: Funding was provided by NRF of South Africa (grant number 142142) and SA MRC (self-initiated research (SIR) grant), and Balvi Foundation. The content and findings reported and illustrated are the sole deduction, view and responsibility of the researchers and do not reflect the official position and sentiments of the funders.
Availability of Data and Materials
All data are available either in Supplementary Information or on request from the first author.
Supplementary Materials
The following supporting information can be downloaded at: https://ojs.exploverpub.com/index.php/jecacm/article/view/201/sup. Movies showing a kinetic series of images for samples treated with either PBS (Supplementary Figure 1) or 28 ng/μL nattokinase (Supplementary Figure 2). Frames start from read 1 at time zero and end at read 9 at time 5 h 28 min. Movies play at 0.5 frames per second. Time zero is the start of the reaction when PBS/NK was added to the sample containing fibrinogen, LPS, Thrombin, and ThT as described in the text. Green annuli are an artefact that may be ignored.
References
- Nagareddy P, Smyth SS. Inflammation and thrombosis in cardiovascular disease. Current Opinion in Hematology 2013; 20(5): 457-463.
- Stark K, Massberg S. Interplay between inflammation and thrombosis in cardiovascular pathology. Nature Reviews Cardiology 2021; 18(9): 666-682.
- Wagner DD, Heger LA. Thromboinflammation: From Atherosclerosis to COVID-19. Arteriosclerosis, Thrombosis, and Vascular Biology 2022; 42(9): 1103-1112.
- leinbongard P, Heusch G. A fresh look at coronary microembolization. Nature Reviews Cardiology 2022; 19(4): 265-280.
- Lacey MJ, Raza S, Rehman H, et al. Coronary Embolism: A Systematic Review. Cardiovascular Revascularization Medicine 2020; 21(3): 367-374.
- Nappi F, Nappi P, Gambardella I, et al. Thromboembolic Disease and Cardiac Thrombotic Complication in COVID-19: A Systematic Review. Metabolites 2022; 12(10): 889.
- Mackman N, Bergmeier W, Stouffer GA, et al. Therapeutic strategies for thrombosis: new targets and approaches. Nature Reviews Drug Discovery 2020; 19(5): 333-352.
- Thakur M, Junho CVC, Bernhard SM, et al. NETs-Induced Thrombosis Impacts on Cardiovascular and Chronic Kidney Disease. Circulation Research 2023; 132(8): 933-949.
- Lippi G, Mattiuzzi C, Favaloro EJ. Novel and emerging therapies: thrombus-targeted fibrinolysis. Seminars in Thrombosis and Hemostasis 2013; 39(1): 48-58.
- Jackson SP, Darbousset R, Schoenwaelder SM. Thromboinflammation: challenges of therapeutically targeting coagulation and other host defense mechanisms. Blood 2019; 133(9): 906-918.
- Medi C, Hankey GJ, Freedman SB. Stroke risk and antithrombotic strategies in atrial fibrillation. Stroke 2010; 41(11): 2705-2713.
- Bikdeli B, Madhavan MV, Jimenez D, et al. COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-up. Journal of the American College of Cardiology 2020; 75(23): 2950-2973.
- Grobler C, Maphumulo SC, Grobbelaar LM, et al. COVID-19: The Rollercoaster of Fibrin(ogen), D-dimer, von Willebrand Factor, P-selectin and Their Interactions with Endothelial Cells, Platelets and Erythrocytes. International Journal of Molecular Sciences 2020; 21(14): 5168.
- Greer IA, Brenner B, Gris JC. Antithrombotic treatment for pregnancy complications: which path for the journey to precision medicine? British Journal of Haematology 2014; 165(5): 585-599.
- Undas A, Brummel-Ziedins K, Mann KG. Why does aspirin decrease the risk of venous thromboembolism? On old and novel antithrombotic effects of acetyl salicylic acid. Journal of Thrombosis and Haemostasis 2014; 12(11): 1776-1787.
- Wang C, Chen J, Tian W, et al. Natto: A medicinal and edible food with health function. Chinese Herbal Medicines 2023; 15(3): 349-359.
- Lampe BJ, English JC. Toxicological assessment of nattokinase derived from Bacillus subtilis var. natto. Food and Chemical Toxicology 2016; 88: 87-99.
- Ruiz Sella SRB, Bueno T, de Oliveira AAB, et al. Bacillus subtilis natto as a potential probiotic in animal nutrition. Critical Reviews in Biotechnology 2021; 41(3): 355-369.
- Afzaal M, Saeed F, Islam F, et al. Nutritional Health Perspective of Natto: A Critical Review. Biochemical Research International 2022; 2022: 5863887.
- Teramoto N, Sato K, Wada T, et al. Impacts of Bacillus subtilis var. natto on the lifespan and stress resistance of Caenorhabditis elegans. Journal of Applied Microbiology 2023; 134(4): lxad082.
- Sumi H, Hamada H, Tsushima H, et al. A novel fibrinolytic enzyme (nattokinase) in the vegetable cheese Natto; a typical and popular soybean food in the Japanese diet. Experientia 1987; 43(10): 1110-1111.
- Sawamura S. On the micro-organisms of natto. The Bulletin of the College of Agriculture, Tokyo Imperial University 1906; 7: 107-110.
- Oshima K. The properties of protease A, the proteolytic enzyme of natto bacteria. Journal of the Society of Agricultural and Forest Chemistry of Sapporo 1925; 71: 387-403.
- Fujita M, Hong K, Ito Y, et al. Transport of nattokinase across the rat intestinal tract. Biological and Pharmaceutical Bulletin 1995; 18(9): 1194-1196.
- Sumi H, Hamada H, Nakanishi K, et al. Enhancement of the fibrinolytic activity in plasma by oral administration of nattokinase. Acta Haematologica 1990; 84(3): 139-143.
- Sumi H, Yanagisawa Y, Yatagai C, et al. Natto bacillus as an oral fibrinolytic agent: Nattokinase activity and the ingestion effect of Bacillus subtilis natto. Food Science and Technology Research 2004; 10(1): 17-20.
- Chen H, McGowan EM, Ren N, et al. Nattokinase: A Promising Alternative in Prevention and Treatment of Cardiovascular Diseases. Biomarker Insights 2018; 13: 1177271918785130.
- Ero MP, Ng CM, Mihailovski T, et al. A pilot study on the serum pharmacokinetics of nattokinase in humans following a single, oral, daily dose. Alternative Therapies in Health and Medicine 2013; 19(3): 16-19.
- Dabbagh F, Negahdaripour M, Berenjian A, et al. Nattokinase: production and application. Applied Microbiology and Biotechnology 2014; 98(22): 9199-9206.
- Kapoor R, Harde H, Jain S, et al. Downstream Processing, Formulation Development and Antithrombotic Evaluation of Microbial Nattokinase. Journal of Biomedical Nanotechnology 2015; 11(7): 1213-1224.
- Zhou XQ, Liu LZ, Zeng XR. Research progress on the utilisation of embedding technology and suitable delivery systems for improving the bioavailability of nattokinase: A review. Food Structure 2021; 30: 100219.
- Liu S, Zhu J, Liu C, et al. Synthesis of sustained release/controlled release nanoparticles carrying nattokinase and their application in thrombolysis. Pharmazie 2021; 76(4): 145-149.
- Priya V, Samridhi, Singh N, et al. Nattokinase Encapsulated Nanomedicine for Targeted Thrombolysis: Development, Improved in Vivo Thrombolytic Effects, and Ultrasound/Photoacoustic Imaging. Molecular Pharmaceutics 2024; 21(1): 283-302.
- Wei X, Luo M, Xie Y, et al. Strain screening, fermentation, separation, and encapsulation for production of nattokinase functional food. Applied Biochemistry and Biotechnology 2012; 168(7): 1753-1764.
- Peng Y, Yang X, Zhang Y. Microbial fibrinolytic enzymes: an overview of source, production, properties, and thrombolytic activity in vivo. Applied Microbiology and Biotechnology 2005; 69(2): 126-132.
- Li D, Hou L, Hu M, et al. Recent Advances in Nattokinase-Enriched Fermented Soybean Foods: A Review. Foods 2022; 11(13): 1867.
- Kurosawa Y, Nirengi S, Homma T, et al. A single-dose of oral nattokinase potentiates thrombolysis and anti-coagulation profiles. Scientific Reports 2015; 5: 11601.
- Selvarajan E, Bhatnagar N. Nattokinase: an updated critical review on challenges and perspectives. Cardiovascular and Hematological Agents in Medicinal Chemistry 2017; 15: 126-135.
- Weng Y, Yao J, Sparks S, et al. Nattokinase: An Oral Antithrombotic Agent for the Prevention of Cardiovascular Disease. International Journal of Molecular Sciences 2017; 18(3): 523.
- Derosa G, Maffioli P, D'Angelo A, et al. Nutraceutical Approach to Preventing Coronavirus Disease 2019 and Related Complications. Frontiers in Immunology 2021; 12: 582556.
- Takagaki S, Suzuki M, Suzuki E, et al. Unsaturated fatty acids enhance the fibrinolytic activity of subtilisin NAT (nattokinase). Journal of Food Biochemistry 2020; 44(8): e13326.
- Li XM, Long JZ, Gao Q, et al. Nattokinase Supplementation and Cardiovascular Risk Factors: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Reviews in Cardiovascular Medicine 2023; 24: 234.
- Jamali N, Vahedi F, Fard ES, et al. Nattokinase: Structure, applications and sources. Biocatalysis and Agricultural Biotechnology 2023; 47: 102564.
- Yuan L, Liangqi C, Xiyu T, et al. Biotechnology, Bioengineering and Applications of Bacillus Nattokinase. Biomolecules 2022; 12(7): 980.
- Nara N, Kurosawa Y, Fuse-Hamaoka S, et al. A single dose of oral nattokinase accelerates skin temperature recovery after cold water immersion: A double-blind, placebo-controlled crossover study. Heliyon 2023; 9(7): e17951.
- Di Micco P, Bernardi FF, Camporese G, et al. Nattokinase historical sketch on experimental and clinical evidence. Italian Journal of Medicine 2023; 17: 1583.
- Kawamata T, Wakimoto A, Nishikawa T, et al. Natto consumption suppresses atherosclerotic plaque progression in LDL receptor-deficient mice transplanted with iRFP-expressing hematopoietic cells. Scientific Reports 2023; 13(1): 22469.
- Sun R, Li D, Sun M, et al. Bacillus natto ameliorates obesity by regulating PI3K/AKT pathways in rats. Biochemical and Biophysical Research Communications 2022; 603: 160-166.
- Ibe S, Kumada K, Yoshida K, et al. Natto (fermented soybean) extract extends the adult lifespan of Caenorhabditis elegans. Bioscience, Biotechnology, and Biochemistry 2013; 77(2): 392-394.
- Guilherme do Prado F, Pagnoncelli MGB, de Melo Pereira GV, et al. Fermented Soy Products and Their Potential Health Benefits: A Review. Microorganisms 2022; 10(8): 1606.
- Yatagai C, Maruyama M, Kawahara T, et al. Nattokinase-promoted tissue plasminogen activator release from human cells. Pathophysiology of Haemostasis and Thrombosis 2008; 36(5): 227-232.
- Takabayashi T, Imoto Y, Sakashita M, et al. Nattokinase, profibrinolytic enzyme, effectively shrinks the nasal polyp tissue and decreases viscosity of mucus. Allergology International 2017; 66(4): 594-602.
- Hsia CH, Shen MC, Lin JS, et al. Nattokinase decreases plasma levels of fibrinogen, factor VII, and factor VIII in human subjects. Nutrition Research 2009; 29(3): 190-196.
- Wang P, Peng C, Xie X, et al. Research progress on the fibrinolytic enzymes produced from traditional fermented foods. Food Science & Nutrition 2023; 11(10): 5675-5688.
- Kumar SS, Sabu A. Fibrinolytic Enzymes for Thrombolytic Therapy. Advances in Experimental Medicine and Biology 2019; 1148: 345-381.
- Fang M, Yuan B, Wang M, et al. Nattokinase: Insights into Biological Activity, Therapeutic Applications, and the Influence of Microbial Fermentation. Fermentation 2023; 9: 950.
- Hazare C, Bhagwat P, Singh S, et al. Diverse origins of fibrinolytic enzymes: A comprehensive review. Heliyon 2024; 10(5): e26668.
- Diwan D, Usmani Z, Sharma M, et al. Thrombolytic Enzymes of Microbial Origin: A Review. International Journal of Molecular Sciences 2021; 22(19): 10468.
- Kotb E. Activity assessment of microbial fibrinolytic enzymes. Applied Microbiology and Biotechnology 2013; 97(15): 6647-6665.
- Mousavi Ghahfarrokhi SS, Mahdigholi FS, Amin M. Collateral beauty in the damages: an overview of cosmetics and therapeutic applications of microbial proteases. Archives of Microbiology 2023; 205(12): 375.
- Liu X, Zeng X, Mahe J, et al. The Effect of Nattokinase-Monascus Supplements on Dyslipidemia: A Four-Month Randomized, Double-Blind, Placebo-Controlled Clinical Trial. Nutrients 2023; 15(19): 4239.
- Huang Z, Chu WK, Ng TK, et al. Protective effects of nattokinase against microvasculopathy and neuroinflammation in diabetic retinopathy. Journal of Diabetes 2023; 15(10): 866-880.
- Liu M, Xu Z, Wang Z, et al. Lipid-lowering, antihypertensive, and antithrombotic effects of nattokinase combined with red yeast rice in patients with stable coronary artery disease: a randomized, double-blinded, placebo-controlled trial. Frontiers in Nutrition 2024; 11: 1380727.
- Wu H, Wang H, Xu F, et al. Acute toxicity and genotoxicity evaluations of Nattokinase, a promising agent for cardiovascular diseases prevention. Regulatory Toxicology and Pharmacology 2019; 103: 205-209.
- Jensen GS, Lenninger M, Ero MP, et al. Consumption of nattokinase is associated with reduced blood pressure and von Willebrand factor, a cardiovascular risk marker: results from a randomized, double-blind, placebo-controlled, multicenter North American clinical trial. Integrated Blood Pressure Control 2016; 9: 95-104.
- Nagata C, Wada K, Tamura T, et al. Dietary soy and natto intake and cardiovascular disease mortality in Japanese adults: the Takayama study. American Journal of Clinical Nutrition 2017; 105(2): 426-431.
- Cai D, Zhu C, Chen S. Microbial production of nattokinase: current progress, challenge and prospect. World Journal of Microbiology and Biotechnology 2017; 33(5): 84.
- Chen PT, Chiang CJ, Chao YP. Medium optimization for the production of recombinant nattokinase by Bacillus subtilis using response surface methodology. Biotechnology Progress 2007; 23(6): 1327-1332.
- Chen H, Chen J, Zhang F, et al. Effective management of atherosclerosis progress and hyperlipidemia with nattokinase: A clinical study with 1,062 participants. Frontiers in Cardiovascular Medicine 2022; 9: 964977.
- Huang Z, Ng TK, Chen W, et al. Nattokinase Attenuates Retinal Neovascularization Via Modulation of Nrf2/HO-1 and Glial Activation. Investigative Ophthalmology & Visual Science 2021; 62(6): 25.
- Lee BH, Lai YS, Wu SC. Antioxidation, angiotensin converting enzyme inhibition activity, nattokinase, and antihypertension of Bacillus subtilis (natto)-fermented pigeon pea. Journal of Food and Drug Analysis 2015; 23(4): 750-757.
- Liu Z, Zhao H, Han L, et al. Improvement of the acid resistance, catalytic efficiency, and thermostability of nattokinase by multisite-directed mutagenesis. Biotechnology and Bioengineering 2019; 116(8): 1833-1843.
- Pan X, Liang P, Teng L, et al. Study on molecular mechanisms of nattokinase in pharmacological action based on label-free liquid chromatography-tandem mass spectrometry. Food Science & Nutrition 2019; 7(10): 3185-3193.
- Tian M, Ning C, Peng S, et al. High-Efficiency Fermentation of Nattokinase by Recombinant PSP2 Using Oyster Protein Hydrolysate as a Substrate. Foods 2023; 12(6): 1252.
- Vianney YM, Tjoa SEE, Aditama R, et al. Designing a less immunogenic nattokinase from Bacillus subtilis subsp. natto: a computational mutagenesis. Journal of Molecular Modeling 2019; 25(11): 337.
- Wu H, Wang H, Li W, et al. Nattokinase-heparin exhibits beneficial efficacy and safety-an optimal strategy for CKD patients on hemodialysis. Glycoconjugate Journal 2019; 36(2): 93-101.
- Wu H, Wang Y, Zhang Y, et al. Breaking the vicious loop between inflammation, oxidative stress and coagulation, a novel anti-thrombus insight of nattokinase by inhibiting LPS-induced inflammation and oxidative stress. Redox Biology 2020; 32: 101500.
- Wu H, Zhang Q, Xu P, et al. Nattokinase Promotes Post-stroke Neurogenesis and Cognition Recovery via Increasing Circulating Irisin. Journal of Agricultural and Food Chemistry 2023; 71(30): 11418-11428.
- Zhang X, Tong Y, Wang J, et al. Screening of a Bacillus subtilis strain producing both nattokinase and milk-clotting enzyme and its application in fermented milk with thrombolytic activity. Journal of Dairy Science 2021; 104(9): 9437-9449.
- Yarnell E. Herbs for Atrial Fibrillation. Alternative and Complementary Therapies 2017; 23: 102-111.
- Gallelli G, Di Mizio G, Palleria C, et al. Data Recorded in Real Life Support the Safety of Nattokinase in Patients with Vascular Diseases. Nutrients 2021; 13(6): 2031.
- Bresson J-L, Burlingame B, Dean T, et al. Safety of fermented soybean extract NSK-SD®as anovel food pursuant to Regulation (EC) No 258/97. EFSA Journal 2016; 14: 4541.
- Yanagisawa Y, Chatake T, Chiba-Kamoshida K, et al. Purification, crystallization and preliminary X-ray diffraction experiment of nattokinase from Bacillus subtilis natto. Acta Crystallographica Section F: Structural Biology and Crystal Communications 2010; 66(Pt 12): 1670-1673.
- Yanagisawa Y, Chatake T, Naito S, et al. X-ray structure determination and deuteration of nattokinase. Journal of Synchrotron Radiation 2013; 20(Pt 6): 875-879.
- Minh NH, Trang HTQ, Van TB, et al. Production and purification of nattokinase from Bacillus subtilis. Food Biotechnology 2022; 36(1): 1-21.
- Wang C, Du M, Zheng D, et al. Purification and characterization of nattokinase from Bacillus subtilis natto B-12. Journal of Agricultural and Food Chemistry 2009; 57(20): 9722-9729.
- Fujita M, Nomura K, Hong K, et al. Purification and characterization of a strong fibrinolytic enzyme (nattokinase) in the vegetable cheese natto, a popular soybean fermented food in Japan. Biochemical and Biophysical Research Communications 1993; 197(3): 1340-1347.
- Yin LJ, Lin HH, Jiang ST. Bioproperties of potent nattokinase from Bacillus subtilis YJ1. Journal of Agricultural and Food Chemistry 2010; 58(9): 5737-5742.
- Pagnoncelli MGB, Fernandes MJ, Rodrigues C, et al. Nattokinases. In Current Developments in Biotechnology and Bioengineering: Production, Isolation and Purification of Industrial Products; Elsevier: Amsterdam, The Netherlands, 2017; pp. 509-526.
- Modi A, Raval I, Doshi P, et al. Heterologous expression of recombinant nattokinase in Escherichia coli BL21(DE3) and media optimization for overproduction of nattokinase using RSM. Protein Expression and Purification 2023; 203: 106198.
- Ni H, Guo PC, Jiang WL, et al. Expression of nattokinase in Escherichia coli and renaturation of its inclusion body. Journal of Biotechnology 2016; 231: 65-71.
- Liang X, Jia S, Sun Y, et al. Secretory expression of nattokinase from Bacillus subtilis YF38 in Escherichia coli. Molecular Biotechnology 2007; 37(3): 187-194.
- Liang X, Zhang L, Zhong J, et al. Secretory expression of a heterologous nattokinase in Lactococcus lactis. Applied Microbiology and Biotechnology 2007; 75(1): 95-101.
- Chen PT, Chiang CJ, Chao YP. Strategy to approach stable production of recombinant nattokinase in Bacillus subtilis. Biotechnology Progress 2007; 23(4): 808-813.
- Liu Z, Zheng W, Ge C, et al. High-level extracellular production of recombinant nattokinase in Bacillus subtilis WB800 by multiple tandem promoters. BMC Microbiology 2019; 19(1): 89.
- Li C, Du Z, Qi S, et al. Food-grade expression of nattokinase in Lactobacillus delbrueckii subsp. bulgaricus and its thrombolytic activity in vitro. Biotechnology Letters 2020; 42(11): 2179-2187.
- Wei X, Zhou Y, Chen J, et al. Efficient expression of nattokinase in Bacillus licheniformis: host strain construction and signal peptide optimization. Journal of Industrial Microbiology and Biotechnology 2015; 42(2): 287-295.
- Guangbo Y, Min S, Wei S, et al. Heterologous expression of nattokinase from B. subtilis natto using Pichia pastoris GS115 and assessment of its thrombolytic activity. BMC Biotechnology 2021; 21(1): 49.
- Sheng Y, Yang J, Wang C, et al. Microbial nattokinase: from synthesis to potential application. Food Function 2023; 14(6): 2568-2585.
- Chen L, Yu K, Ma A, et al. Enhanced Thermostability of Nattokinase by Computation-Based Rational Redesign of Flexible Regions. Journal of Agricultural and Food Chemistry 2024; 72(25): 14241-14254.
- Urano T, Ihara H, Umemura K, et al. The profibrinolytic enzyme subtilisin NAT purified from Bacillus subtilis cleaves and inactivates plasminogen activator inhibitor type 1. Journal of Biological Chemistry 2001; 276(27): 24690-24696.
- Jang JY, Kim TS, Cai J, et al. Nattokinase improves blood flow by inhibiting platelet aggregation and thrombus formation. Laboratory Animal Research 2013; 29(4): 221-225.
- Fujita M, Ohnishi K, Takaoka S, et al. Antihypertensive effects of continuous oral administration of nattokinase and its fragments in spontaneously hypertensive rats. Biological and Pharmaceutical Bulletin 2011; 34(11): 1696-1701.
- Kim JY, Gum SN, Paik JK, et al. Effects of nattokinase on blood pressure: a randomized, controlled trial. Hypertension Research 2008; 31(8): 1583-1588.
- Elbakry MMM, Mansour SZ, Helal H, et al. Nattokinase attenuates bisphenol A or gamma irradiation-mediated hepatic and neural toxicity by activation of Nrf2 and suppression of inflammatory mediators in rats. Environmental Science and Pollution Research International 2022; 29(49): 75086-75100.
- Maslarov D, Drenska D, Maslarova-Gelov J, et al. Understanding the concept of Nattokinase use: a few years after beginning. Biotechnology and Biotechnological Equipment 2023; 37: 2249552.
- Pretorius E, Swanepoel AC, Oberholzer HM, et al. A descriptive investigation of the ultrastructure of fibrin networks in thrombo-embolic ischemic stroke. Journal of Thrombosis and Thrombolysis 2011; 31(4): 507-513.
- Pretorius E. The use of a desktop scanning electron microscope as a diagnostic tool in studying fibrin networks of thrombo-embolic ischemic stroke. Ultrastructural Pathology 2011; 35(6): 245-250.
- Pretorius E, Oberholzer HM, van der Spuy WJ, et al. Qualitative scanning electron microscopy analysis of fibrin networks and platelet abnormalities in diabetes. Blood Coagulation & Fibrinolysis 2011; 22(6): 463-467.
- Pretorius E, Vermeulen N, Bester J, et al. A novel method for assessing the role of iron and its functional chelation in fibrin fibril formation: the use of scanning electron microscopy. Toxicology Mechanisms and Methods 2013; 23(5): 352-359.
- Pretorius E, Mbotwe S, Bester J, et al. Acute induction of anomalous and amyloidogenic blood clotting by molecular amplification of highly substoichiometric levels of bacterial lipopolysaccharide. Journal of the Royal Society Interface 2016; 123(13): 20160539.
- Kell DB, Pretorius E. Proteins behaving badly. Substoichiometric molecular control and amplification of the initiation and nature of amyloid fibril formation: lessons from and for blood clotting. Progress in Biophysics and Molecular Biology 2017; 123: 16-41.
- Pretorius E, Mbotwe S, Kell DB. Lipopolysaccharide-binding protein (LBP) reverses the amyloid state of fibrin seen in plasma of type 2 diabetics with cardiovascular comorbidities. Scientific Reports 2017; 7: 9680.
- Pretorius E, Page MJ, Hendricks L, et al. Both lipopolysaccharide and lipoteichoic acids potently induce anomalous fibrin amyloid formation: assessment with novel AmytrackerTM stains. bioRxiv 2017.
- Pretorius E, Page MJ, Engelbrecht L, et al. Substantial fibrin amyloidogenesis in type 2 diabetes assessed using amyloid-selective fluorescent stains. Cardiovascular Diabetology 2017; 16: 141.
- Pretorius E, Page MJ, Mbotwe S, et al. Lipopolysaccharide-binding protein (LBP) can reverse the amyloid state of fibrin seen or induced in Parkinson’s disease. PLoS One 2018; 13(3): e0192121.
- Pretorius E, Page MJ, Hendricks L, et al. Both lipopolysaccharide and lipoteichoic acids potently induce anomalous fibrin amyloid formation: assessment with novel AmytrackerTM stains. Journal of the Royal Society Interface 2018; 15(139): 20170941.
- Pretorius E, Bester J, Page MJ, et al. The potential of LPS-binding protein to reverse amyloid formation in plasma fibrin of individuals with Alzheimer-type dementia. Frontiers in Aging Neuroscience 2018; 10: 257.
- de Waal GM, Engelbrecht L, Davis T, et al. Correlative Light-Electron Microscopy detects lipopolysaccharide and its association with fibrin fibres in Parkinson's Disease, Alzheimer's Disease and Type 2 Diabetes Mellitus. Scientific Reports 2018; 8(1): 16798.
- Pretorius E, Venter C, Laubscher GJ, et al. Prevalence of symptoms, comorbidities, fibrin amyloid microclots and platelet pathology in individuals with Long COVID/Post-Acute Sequelae of COVID-19 (PASC). Cardiovascular Diabetology 2022; 21: 148.
- Kell DB, Pretorius E. Are fibrinaloid microclots a cause of autoimmunity in Long Covid and other post-infection diseases? Biochemical Journal 2023; 480: 1217-1240.
- Candelise N, Baiardi S, Franceschini A, et al. Towards an improved early diagnosis of neurodegenerative diseases: the emerging role of in vitro conversion assays for protein amyloids. Acta Neuropathologica Communications 2020; 8(1): 117.
- Poleggi A, Baiardi S, Ladogana A, et al. The Use of Real-Time Quaking-Induced Conversion for the Diagnosis of Human Prion Diseases. Frontiers in Aging Neuroscience 2022; 14: 874734.
- Grobbelaar LM, Venter C, Vlok M, et al. SARS-CoV-2 spike protein S1 induces fibrin(ogen) resistant to fibrinolysis: implications for microclot formation in COVID-19. Bioscience Reports 2021; 41(8): BSR20210611.
- Kell DB, Pretorius E. The simultaneous occurrence of both hypercoagulability and hypofibrinolysis in blood and serum during systemic inflammation, and the roles of iron and fibrin(ogen). Integrative Biology 2015; 7: 24-52.
- Bergaglio T, Synhaivska O, Nirmalraj PN. 3D Holo-tomographic Mapping of COVID-19 Microclots in Blood to Assess Disease Severity. ACS Chemical Biology and Biomedical Imaging 2024; 2(3): 194-204.
- Stewart KL, Radford SE. Amyloid plaques beyond Abeta: a survey of the diverse modulators of amyloid aggregation. Biophysical Reviews 2017; 9: 405-419.
- Gligorijević N, Simeon M, Mirjana R, et al. Ligand binding to fibrinogen influences its structure and function. Biologia Serbica 2021; 43: 24-31.
- Gligorijević N, Vasović T, Lević S, et al. Atypical antipsychotic clozapine binds fibrinogen and affects fibrin formation. International Journal of Biological Macromolecules 2020; 154: 142-149.
- Liu B, Moloney A, Meehan S, et al. Iron promotes the toxicity of amyloid beta peptide by impeding its ordered aggregation. Journal of Biological Chemistry 2011; 286(6): 4248-4256.
- Louros N, Schymkowitz J, Rousseau F. Mechanisms and pathology of protein misfolding and aggregation. Nature Reviews Molecular Cell Biology 2023; 24(12): 912-933.
- Xu Y, Maya-Martinez R, Guthertz N, et al. Tuning the rate of aggregation of hIAPP into amyloid using small-molecule modulators of assembly. Nature Communications 2022; 13(1): 1040.
- Heller GT, Aprile FA, Michaels TCT, et al. Small-molecule sequestration of amyloid-beta as a drug discovery strategy for Alzheimer's disease. Science Advances 2020; 6(45): eabb5924.
- Ziaunys M, Sneideris T, Smirnovas V. Formation of distinct prion protein amyloid fibrils under identical experimental conditions. Scientific Reports 2020; 10(1): 4572.
- Ziaunys M, Sakalauskas A, Mikalauskaite K, et al. Temperature-Dependent Structural Variability of Prion Protein Amyloid Fibrils. International Journal of Molecular Sciences 2021; 22(10): 5075.
- Watts JC, Condello C, Stohr J, et al. Serial propagation of distinct strains of Abeta prions from Alzheimer's disease patients. Proceedings of the National Academy of Sciences of the United States of America 2014; 111(28): 10323-10328.
- Han ZZ, Kang SG, Arce L, et al. Prion-like strain effects in tauopathies. Cell and Tissue Research 2023; 392(1): 179-199.
- Shoup D, Priola SA. Cell biology of prion strains in vivo and in vitro. Cell and Tissue Research 2023; 392(1): 269-283.
- Bartz JC. Prion Strain Diversity. Cold Spring Harbor Perspectives in Medicine 2016; 6(12): a024349.
- Igel-Egalon A, Béringue V, Rezaei H, et al. Prion Strains and Transmission Barrier Phenomena. Pathogens 2018; 7(1): 5.
- Scialò C, De Cecco E, Manganotti P, et al. Prion and Prion-Like Protein Strains: Deciphering the Molecular Basis of Heterogeneity in Neurodegeneration. Viruses 2019; 11(3): 261.
- Levavasseur E, Privat N, Haïk S. In vitro Modeling of Prion Strain Tropism. Viruses 2019; 11(3): 236.
- Peden AH, Suleiman S, Barria MA. Understanding Intra-Species and Inter-Species Prion Conversion and Zoonotic Potential Using Protein Misfolding Cyclic Amplification. Frontiers in Aging Neuroscience 2021; 13: 716452.
- Hoyt F, Alam P, Artikis E, et al. Cryo-EM of prion strains from the same genotype of host identifies conformational determinants. PLoS Pathogens 2022; 18(11): e1010947.
- Igel A, Fornara B, Rezaei H, et al. Prion assemblies: structural heterogeneity, mechanisms of formation, and role in species barrier. Cell and Tissue Research 2023; 392(1): 149-166.
- Sharma A, Bruce KL, Chen B, et al. Contributions of the Prion Protein Sequence, Strain, and Environment to the Species Barrier. Journal of Biological Chemistry 2016; 291(3): 1277-1288.
- Fändrich M, Meinhardt J, Grigorieff N. Structural polymorphism of Alzheimer Abeta and other amyloid fibrils. Prion 2009; 3(2): 89-93.
- Kodali R, Williams AD, Chemuru S, et al. Abeta(1-40) forms five distinct amyloid structures whose beta-sheet contents and fibril stabilities are correlated. Journal of Molecular Biology 2010; 401(3): 503-517.
- Fändrich M, Wulff M, Pedersen JS, et al. Fibrillar Polymorphism. In Amyloid Fibrils and Prefibrillar Aggregates: Molecular and Biological Properties; Wiley-VCH: Weinheim, Germany, 2013; pp. 321–343.
- Tycko R. Physical and structural basis for polymorphism in amyloid fibrils. Protein Science 2014; 23(11): 1528-1539.
- Lutter L, Serpell CJ, Tuite MF, et al. The molecular lifecycle of amyloid—Mechanism of assembly, mesoscopic organisation, polymorphism, suprastructures, and biological consequences. Biochimica et Biophysica Acta - Proteins and Proteomics 2019; 1867(11): 140257.
- Arifin MI, Hannaoui S, Chang SC, et al. Cervid Prion Protein Polymorphisms: Role in Chronic Wasting Disease Pathogenesis. International Journal of Molecular Sciences 2021; 22(5): 2271.
- Farzadfard A, Kunka A, Mason TO, et al. Thermodynamic characterization of amyloid polymorphism by microfluidic transient incomplete separation. Chemical Science 2024; 15(7): 2528-2544.
- Pfeiffer PB, Ugrina M, Schwierz N, et al. Cryo-EM Analysis of the Effect of Seeding with Brain-derived Abeta Amyloid Fibrils. Journal of Molecular Biology 2024; 436(4): 168422.
- Mishra S. Emerging Trends in Cryo-EM-based Structural Studies of Neuropathological Amyloids. Journal of Molecular Biology 2023; 435(24): 168361.
- Petkova AT, Leapman RD, Guo Z, et al. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science 2005; 307(5707): 262-265.
- Peccati F, Pantaleone S, Solans-Monfort X, et al. Fluorescent Markers for Amyloid-beta Detection: Computational Insights. Israel Journal of Chemistry 2017; 57: 686-698.
- Peccati F, Pantaleone S, Riffet V, et al. Binding of Thioflavin T and Related Probes to Polymorphic Models of Amyloid-beta Fibrils. Journal of Physical Chemistry B 2017; 121(38): 8926-8934.
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nature Reviews Genetics 2005; 6(6): 435-450.
- Wiltzius JJW, Landau M, Nelson R, et al. Molecular mechanisms for protein-encoded inheritance. Nature Structural & Molecular Biology 2009; 16(9): 973-978.
- Liebman SW, Chernoff YO. Prions in yeast. Genetics 2012; 191(4): 1041-1072.
- Wickner RB, Shewmaker FP, Bateman DA, et al. Yeast prions: structure, biology, and prion-handling systems. Microbiology and Molecular Biology Reviews 2015; 79(1): 1-17.
- Wickner RB, Edskes HK, Gorkovskiy A, et al. Yeast and Fungal Prions: Amyloid-Handling Systems, Amyloid Structure, and Prion Biology. Advanced Genetics 2016; 93: 191-236.
- Bao J, Wen Z, Kim M, et al. Identifying highly heritable brain amyloid phenotypes through mining Alzheimer's imaging and sequencing biobank data. Pacific Symposium on Biocomputing 2022; 27: 109-120.
- Telling GC. The shape of things to come: structural insights into how prion proteins encipher heritable information. Nature Communications 2022; 13(1): 4003.
- Jarrett JT, Lansbury PT, Jr. Seeding "one-dimensional crystallization" of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell 1993; 73(6): 1055-1058.
- Luers L, Bannach O, Stöhr J, et al. Seeded fibrillation as molecular basis of the species barrier in human prion diseases. PLoS One 2013; 8(8): e72623.
- Pinotsi D, Michel CH, Buell AK, et al. Nanoscopic insights into seeding mechanisms and toxicity of alpha-synuclein species in neurons. Proceedings of the National Academy of Sciences of the United States of America 2016; 113(14): 3815-3819.
- Kaufman SK, Thomas TL, Del Tredici K, et al. Characterization of tau prion seeding activity and strains from formaldehyde-fixed tissue. Acta Neuropathologica Communications 2017; 5(1): 41.
- Mudher A, Colin M, Dujardin S, et al. What is the evidence that tau pathology spreads through prion-like propagation? Acta Neuropathologica Communications 2017; 5(1): 99.
- Manca M, Kraus A. Defining the Protein Seeds of Neurodegeneration using Real-Time Quaking-Induced Conversion Assays. Biomolecules 2020; 10(9): 1233.
- Peng C, Trojanowski JQ, Lee VM. Protein transmission in neurodegenerative disease. Nature Reviews Neurology 2020; 16(4): 199-212.
- Standke HG, Kraus A. Seed amplification and RT-QuIC assays to investigate protein seed structures and strains. Cell and Tissue Research 2022; 392(1): 323-335.
- Thacker D, Sanagavarapu K, Frohm B, et al. The role of fibril structure and surface hydrophobicity in secondary nucleation of amyloid fibrils. Proceedings of the National Academy of Sciences of the United States of America 2020; 117(41): 25272-25283.
- Coysh T, Mead S. The Future of Seed Amplification Assays and Clinical Trials. Frontiers in Aging Neuroscience 2022; 14: 872629.
- Vaneyck J, Yousif TA, Segers-Nolten I, et al. Quantitative Seed Amplification Assay: A Proof-of-Principle Study. The Journal of Physical Chemistry B 2023; 127(8): 1735-1743.
- Sulskis D, Sneideriene G, Ziaunys M, et al. The seeding barrier between human and Syrian hamster prion protein amyloid fibrils is determined by beta2-alpha2 loop sequence elements. International Journal of Biological Macromolecules 2023; 238: 124038.
- Scheres SHW, Ryskeldi-Falcon B, Goedert M. Molecular pathology of neurodegenerative diseases by cryo-EM of amyloids. Nature 2023; 621(7980): 701-710.
- Yang Y, Arseni D, Zhang W, et al. Cryo-EM structures of amyloid-beta 42 filaments from human brains. Science 2022; 375(6577): 167-172.
- Heerde T, Rennegarbe M, Biedermann A, et al. Cryo-EM demonstrates the in vitro proliferation of an ex vivo amyloid fibril morphology by seeding. Nature Communications 2022; 13(1): 85.
- Bondarev SA, Antonets KS, Kajava AV, et al. Protein Co-Aggregation Related to Amyloids: Methods of Investigation, Diversity, and Classification. International Journal of Molecular Sciences 2018; 19(8): 2292.
- Sidhu A, Segers-Nolten I, Subramaniam V. Conformational Compatibility Is Essential for Heterologous Aggregation of alpha-Synuclein. ACS Chemical Neuroscience 2016; 7(6): 719-727.
- Ivanova MI, Lin Y, Lee YH, et al. Biophysical processes underlying cross-seeding in amyloid aggregation and implications in amyloid pathology. Biophysical Chemistry 2021; 269: 106507.
- Subedi S, Sasidharan S, Nag N, et al. Amyloid Cross-Seeding: Mechanism, Implication, and Inhibition. Molecules 2022; 27(6): 1776.
- Chatterjee D, Jacob RS, Ray S, et al. Co-aggregation and secondary nucleation in the life cycle of human prolactin/galanin functional amyloids. Elife 2022; 11: e73835.
- Semerdzhiev SA, Segers-Nolten I, van der Schoot P, et al. SARS-CoV-2 N-protein induces the formation of composite alpha-synuclein/N-protein fibrils that transform into a strain of alpha-synuclein fibrils. Nanoscale 2023; 15(45): 18337-18346.
- Willbold D, Strodel B, Schroder GF, et al. Amyloid-type Protein Aggregation and Prion-like Properties of Amyloids. Chemical Reviews 2021; 121(13): 8285-8307.
- Zielinski M, Röder C, Schröder GF. Challenges in sample preparation and structure determination of amyloids by cryo-EM. Journal of Biological Chemistry 2021; 297(2): 100938.
- Pretorius E, Vlok M, Venter C, et al. Persistent clotting protein pathology in Long COVID/ Post-Acute Sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin. Cardiovascular Diabetology 2021; 20: 172.
- Turner S, Khan MA, Putrino D, et al. Long COVID: pathophysiological factors and abnormal coagulation. Trends in Endocrinology & Metabolism 2023; 34: 321-344.
- Turner S, Laubscher GJ, Khan MA, et al. Accelerating discovery: A novel flow cytometric method for detecting fibrin(ogen) amyloid microclots using long COVID as a model. Heliyon 2023; 9: e19605.
- Turner S, Naidoo CA, Usher TJ, et al. Increased Levels of Inflammatory and Endothelial Biomarkers in Blood of Long COVID Patients Point to Thrombotic Endothelialitis. Seminars in Thrombosis and Hemostasis 2024; 50(2): 288-294.
- Dalton CF, de Oliveira MIR, Stafford P, et al. Increased fibrinaloid microclot counts in platelet-poor plasma are associated with Long COVID. medRxiv 2024.
- Okuducu YK, Boribong B, Ellett F, et al. Evidence Circulating Microclots and Activated Platelets Contribute to Hyperinflammation Within Pediatric Post Acute Sequala of COVID. American Journal of Respiratory and Critical Care Medicine 2024; 209: A2247.
- Nunes JM, Kruger A, Proal A, et al. The Occurrence of Hyperactivated Platelets and Fibrinaloid Microclots in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). Pharmaceuticals 2022; 15(8): 931.
- Nunes JM, Kell DB, Pretorius E. Cardiovascular and haematological pathology in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS): a role for Viruses. Blood Reviews 2023; 60: 101075.
- Grobbelaar LM, Kruger A, Venter C, et al. Relative hypercoagulopathy of the SARS-CoV-2 Beta and Delta variants when compared to the less severe Omicron variants is related to TEG parameters, the extent of fibrin amyloid microclots, and the severity of clinical illness. Seminars in Thrombosis and Hemostasis 2022; 48: 858-868.
- Pretorius E, Kell DB. A perspective on how microscopy imaging of fibrinaloid microclots and platelet pathology may be applied in clinical investigations. Seminars in Thrombosis and Hemostasis 2024; 50(4): 537-551.
- Kell DB, Laubscher GJ, Pretorius E. A central role for amyloid fibrin microclots in long COVID/PASC: origins and therapeutic implications. Biochemical Journal 2022; 479: 537-559.
- Kell DB, Pretorius E. The potential role of ischaemia-reperfusion injury in chronic, relapsing diseases such as rheumatoid arthritis, long COVID and ME/CFS: evidence, mechanisms, and therapeutic implications. Biochemical Journal 2022; 479: 1653-1708.
- Kell DB, Khan MA, Kane B, et al. Possible role of fibrinaloid microclots in Postural Orthostatic Tachycardia Syndrome (POTS): focus on Long COVID. Journal of Personalized Medicine 2024; 14: 170.
- Schofield J, Abrams ST, Jenkins R, et al. Amyloid-fibrinogen aggregates ("microclots") predict risks of Disseminated Intravascular Coagulation and mortality. Blood Advances 2024; 8: 2499-2508.
- Appelman B, Charlton BT, Goulding RP, et al. Muscle abnormalities worsen after post-exertional malaise in long COVID. Nature Communications 2024; 15(1): 17.
- Thagard P. Explaining disease: Correlations, causes, and mechanisms. Minds and Machines 1998; 8(1): 61-78.
- Thagard P. How scientists explain disease; Princeton University Press: Princeton, NJ, USA, 1999.
- Thagard P. Coherence, truth, and the development of scientific knowledge. Philosophy of Science 2007; 74(1): 28-47.
- Thagard P. Explanatory Coherence. In Reasoning: Studies of Human Inference and Its Foundations; Cambridge University Press: Cambridge, UK, 2008; pp. 471-513.
- Nelson R, Sawaya MR, Balbirnie M, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature 2005; 435(7043): 773-778.
- Tycko R, Wickner RB. Molecular structures of amyloid and prion fibrils: consensus versus controversy. Accounts of Chemical Research 2013; 46(7): 1487-1496.
- Serpell LC, Sunde M, Benson MD, et al. The protofilament substructure of amyloid fibrils. Journal of Molecular Biology 2000; 300(5): 1033-1039.
- Jahn TR, Makin OS, Morris KL, et al. The common architecture of cross-beta amyloid. Journal of Molecular Biology 2010; 395(4): 717-727.
- Morris KL, Serpell LC. From Molecular to Supramolecular Amyloid Structures: Contributions from Fiber Diffraction and Electron Microscopy. In Amyloid Fibrils and Prefibrillar Aggregates: Molecular and Biological Properties; Wiley-VCH: Weinheim, Germany, 2013; pp. 63-84.
- Iadanza MG, Jackson MP, Hewitt EW, et al. A new era for understanding amyloid structures and disease. Nature Reviews Molecular Cell Biology 2018; 19(12): 755-773.
- Thurber KR, Yau WM, Tycko R. Structure of Amyloid Peptide Ribbons Characterized by Electron Microscopy, Atomic Force Microscopy, and Solid-State Nuclear Magnetic Resonance. The Journal of Physical Chemistry B 2024; 128(7): 1711-1723.
- Gremer L, Scholzel D, Schenk C, et al. Fibril structure of amyloid-beta(1-42) by cryo-electron microscopy. Science 2017; 358(6359): 116-119.
- Chen GF, Xu TH, Yan Y, et al. Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacologica Sinica 2017; 38(9): 1205-1235.
- Gallardo R, Ranson NA, Radford SE. Amyloid structures: much more than just a cross-beta fold. Current Opinion in Structural Biology 2020; 60: 7-16.
- Greenwald J, Riek R. Biology of amyloid: structure, function, and regulation. Structure 2010; 18(10): 1244-1260.
- Fändrich M, Schmidt M, Grigorieff N. Recent progress in understanding Alzheimer's beta-amyloid structures. Trends in Biochemical Sciences 2011; 36(6): 338-345.
- Eisenberg DS, Sawaya MR. Structural Studies of Amyloid Proteins at the Molecular Level. Annual Review of Biochemistry 2017; 86: 69-95.
- Heerde T, Bansal A, Schmidt M, et al. Cryo-EM structure of a catalytic amyloid fibril. Scientific Reports 2023; 13(1): 4070.
- Heerde T, Schütz D, Lin YJ, et al. Cryo-EM structure and polymorphic maturation of a viral transduction enhancing amyloid fibril. Nature Communications 2023; 14(1): 4293.
- Gonay V, Dunne MP, Caceres-Delpiano J, et al. Developing machine-learning-based amyloid predictors with Cross-Beta DB. bioRxiv 2024.
- Bücker R, Seuring C, Cazey C, et al. The Cryo-EM structures of two amphibian antimicrobial cross-beta amyloid fibrils. Nature Communications 2022; 13(1): 4356.
- Khurana R, Coleman C, Ionescu-Zanetti C, et al. Mechanism of thioflavin T binding to amyloid fibrils. Journal of Structural Biology 2005; 151(3): 229-238.
- Hawe A, Sutter M, Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharmaceutical Research 2008; 25(7): 1487-1499.
- Amdursky N, Erez Y, Huppert D. Molecular rotors: what lies behind the high sensitivity of the thioflavin-T fluorescent marker. Accounts of Chemical Research 2012; 45(9): 1548-1557.
- Biancalana M, Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochimica et Biophysica Acta 2010; 1804(7): 1405-1412.
- Gade Malmos K, Blancas-Mejia LM, Weber B, et al. ThT 101: a primer on the use of thioflavin T to investigate amyloid formation. Amyloid 2017; 24(1): 1-16.
- Lee JE, Sang JC, Rodrigues M, et al. Mapping Surface Hydrophobicity of alpha-Synuclein Oligomers at the Nanoscale. Nano Letters 2018; 18(12): 7494-7501.
- Taylor CG, Meisl G, Horrocks MH, et al. Extrinsic Amyloid-Binding Dyes for Detection of Individual Protein Aggregates in Solution. Analytical Chemistry 2018; 90(17): 10385-10393.
- De S, Klenerman D. Imaging individual protein aggregates to follow aggregation and determine the role of aggregates in neurodegenerative disease. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2019; 1867(10): 870-878.
- Sulatskaya AI, Rychkov GN, Sulatsky MI, et al. New Evidence on a Distinction between A beta 40 and A beta 42 Amyloids: Thioflavin T Binding Modes, Clustering Tendency, Degradation Resistance, and Cross-Seeding. International Journal of Molecular Sciences 2022; 23(10): 5513.
- Xue C, Lin TY, Chang D, et al. Thioflavin T as an amyloid dye: fibril quantification, optimal concentration and effect on aggregation. Royal Society Open Science 2017; 4(1): 160696.
- Arad E, Green H, Jelinek R, et al. Revisiting thioflavin T (ThT) fluorescence as a marker of protein fibrillation—The prominent role of electrostatic interactions. Journal of Colloid and Interface Science 2020; 573: 87-95.
- Lucignano R, Spadaccini R, Merlino A, et al. Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials. International Journal of Biological Macromolecules 2024; 254(Pt 1): 127775.
- Pujols J, Peña-Díaz S, Lázaro DF, et al. Small molecule inhibits alpha-synuclein aggregation, disrupts amyloid fibrils, and prevents degeneration of dopaminergic neurons. Proceedings of the National Academy of Sciences of the United States of America 2018; 115(41): 10481-10486.
- Bertoncini CW, Celej MS. Small molecule fluorescent probes for the detection of amyloid self-assembly in vitro and in vivo. Current Protein & Peptide Science 2011; 12(3): 205-220.
- Panda C, Sharma LG, Pandey LM. Experimental procedures to investigate fibrillation of proteins. MethodsX 2023; 11: 102445.
- Choo LP, Wetzel DL, Halliday WC, et al. In situ characterization of beta-amyloid in Alzheimer's diseased tissue by synchrotron Fourier transform infrared microspectroscopy. Biophysical Journal 1996; 71(4): 1672-1679.
- Cerdà-Costa N, De la Arada I, Avilés FX, et al. Influence of aggregation propensity and stability on amyloid fibril formation as studied by Fourier transform infrared spectroscopy and two-dimensional COS analysis. Biochemistry 2009; 48(44): 10582-10590.
- Prater C, Bai Y, Konings SC, et al. Fluorescently Guided Optical Photothermal Infrared Microspectroscopy for Protein-Specific Bioimaging at Subcellular Level. Journal of Medicinal Chemistry 2023; 66(4): 2542-2549.
- Zhang G, Babenko V, Dzwolak W, et al. Dimethyl Sulfoxide Induced Destabilization and Disassembly of Various Structural Variants of Insulin Fibrils Monitored by Vibrational Circular Dichroism. Biochemistry 2015; 54(49): 7193-7202.
- Cornejo A, Aguilar Sandoval F, Caballero L, et al. Rosmarinic acid prevents fibrillization and diminishes vibrational modes associated to beta sheet in tau protein linked to Alzheimer's disease. Journal of Enzyme Inhibition and Medicinal Chemistry 2017; 32(1): 945-953.
- Li S, Luo Z, Zhang R, et al. Distinguishing Amyloid beta-Protein in a Mouse Model of Alzheimer's Disease by Label-Free Vibrational Imaging. Biosensors 2021; 11(10): 365.
- Watson MD, Lee JC. Coupling chemical biology and vibrational spectroscopy for studies of amyloids in vitro and in cells. Current Opinion in Chemical Biology 2021; 64: 90-97.
- Ami D, Natalello A. Characterization of the Conformational Properties of Soluble and Insoluble Proteins by Fourier Transform Infrared Spectroscopy. Methods in Molecular Biology 2022; 2406: 439-454.
- Vu KHP, Blankenburg GH, Lesser-Rojas L, et al. Applications of Single-Molecule Vibrational Spectroscopic Techniques for the Structural Investigation of Amyloid Oligomers. Molecules 2022; 27(19): 6448.
- Ami D, Mereghetti P, Natalello A. Contribution of Infrared Spectroscopy to the Understanding of Amyloid Protein Aggregation in Complex Systems. Frontiers in Molecular Biosciences 2022; 9: 822852.
- de la Arada I, Seiler C, Mantele W. Amyloid fibril formation from human and bovine serum albumin followed by quasi-simultaneous Fourier-transform infrared (FT-IR) spectroscopy and static light scattering (SLS). European Biophysics Journal 2012; 41(11): 931-938.
- Ridgley DM, Claunch EC, Barone JR. Characterization of large amyloid fibers and tapes with Fourier transform infrared (FT-IR) and Raman spectroscopy. Applied Spectroscopy 2013; 67(12): 1417-1426.
- Biancalana M, Makabe K, Koide A, et al. Molecular mechanism of thioflavin-T binding to the surface of beta-rich peptide self-assemblies. Journal of Molecular Biology 2009; 385(4): 1052-1063.
- Sidhu A, Vaneyck J, Blum C, et al. Polymorph-specific distribution of binding sites determines thioflavin-T fluorescence intensity in alpha-synuclein fibrils. Amyloid 2018; 25(3): 189-196.
- Chisholm TS, Hunter CA. A closer look at amyloid ligands, and what they tell us about protein aggregates. Chemical Society Reviews 2024; 53(3): 1354-1374.
- Rovnyagina NR, Sluchanko NN, Tikhonova TN, et al. Binding of thioflavin T by albumins: An underestimated role of protein oligomeric heterogeneity. International Journal of Biological Macromolecules 2018; 108: 284-290.
- Rovnyagina NR, Budylin GS, Vainer YG, et al. Fluorescence Lifetime and Intensity of Thioflavin T as Reporters of Different Fibrillation Stages: Insights Obtained from Fluorescence Up-Conversion and Particle Size Distribution Measurements. International Journal of Molecular Sciences 2020; 21(17): 6169.
- Rovnyagina NR, Tikhonova TN, Kompanets VO, et al. Free and bound Thioflavin T molecules with ultrafast relaxation: implications for assessment of protein binding and aggregation. Laser Physics Letters 2019; 16: 075601.
- Mikalauskaite K, Ziaunys M, Sneideris T, et al. Effect of Ionic Strength on Thioflavin-T Affinity to Amyloid Fibrils and Its Fluorescence Intensity. International Journal of Molecular Sciences 2020; 21(23): 8916.
- Ziaunys M, Sakalauskas A, Mikalauskaite K, et al. Exploring the occurrence of thioflavin-T-positive insulin amyloid aggregation intermediates. PeerJ 2021; 9: e10918.
- Wolfe LS, Calabrese MF, Nath A, et al. Protein-induced photophysical changes to the amyloid indicator dye thioflavin T. Proceedings of the National Academy of Sciences of the United States of America 2010; 107(39): 16863-16868.
- Ziaunys M, Smirnovas V. Additional Thioflavin-T Binding Mode in Insulin Fibril Inner Core Region. The Journal of Physical Chemistry B 2019; 123(41): 8727-8732.
- Ziaunys M, Mikalauskaite K, Sakalauskas A, et al. Investigating lysozyme amyloid fibril formation and structural variability dependence on its initial folding state under different pH conditions. Protein Science 2024; 33(2): e4888.
- Chaari A, Fahy C, Chevillot-Biraud A, et al. Investigating the effects of different natural molecules on the structure and oligomerization propensity of hen egg-white lysozyme. International Journal of Biological Macromolecules 2019; 134: 189-201.
- Anselmo S, Sancataldo G, Vetri V. Deciphering amyloid fibril molecular maturation through FLIM-phasor analysis of thioflavin T. Biophysical Reports 2024; 4(1): 100145.
- Lindberg DJ, Wranne MS, Gilbert Gatty M, et al. Steady-state and time-resolved Thioflavin-T fluorescence can report on morphological differences in amyloid fibrils formed by Abeta(1-40) and Abeta(1-42). Biochemical and Biophysical Research Communications 2015; 458(2): 418-423.
- Sulatskaya AI, Rodina NP, Polyakov DS, et al. Structural Features of Amyloid Fibrils Formed from the Full-Length and Truncated Forms of Beta-2-Microglobulin Probed by Fluorescent Dye Thioflavin T. International Journal of Molecular Sciences 2018; 19(9): 2762.
- De Luca G, Fennema Galparsoro D, Sancataldo G, et al. Probing ensemble polymorphism and single aggregate structural heterogeneity in insulin amyloid self-assembly. Journal of Colloid and Interface Science 2020; 574: 229-240.
- Thompson AJ, Herling TW, Kubankova M, et al. Molecular Rotors Provide Insights into Microscopic Structural Changes During Protein Aggregation. The Journal of Physical Chemistry B 2015; 119(32): 10170-10179.
- Krebs MRH, Bromley EH, Donald AM. The binding of thioflavin-T to amyloid fibrils: localisation and implications. Journal of Structural Biology 2005; 149(1): 30-37.
- Noormägi A, Primar K, Tõugu V, et al. Interference of low-molecular substances with the thioflavin-T fluorescence assay of amyloid fibrils. Journal of Peptide Science 2012; 18(1): 59-64.
- Hulscher N, Procter BC, Wynn C, et al. Clinical Approach to Post-acute Sequelae After COVID-19 Infection and Vaccination. Cureus 2023; 15(11): e49204.
- McCullough PA, Wynn C, Procter BC. Clinical Rationale for SARS-CoV-2 Base Spike Protein Detoxification in Post COVID-19 and Vaccine Injury Syndromes. Journal of the American Physicians and Surgeons 2023; 28: 90-93.
- Yang M, Wu J, Huang Q, et al. Probing the Role of Catalytic Triad on the Cleavage between Intramolecular Chaperone and NK Mature Peptide. Journal of Agricultural and Food Chemistry 2021; 69(7): 2348-2353.
- Fujita M, Ito Y, Hong K, et al. Characterization of Nattokinase-degraded Products from Human Fibrinogen or Cross-linked Fibrin. Fibrinolysis 1995; 9: 157-164.
- Tanikawa T, Kiba Y, Yu J, et al. Degradative Effect of Nattokinase on Spike Protein of SARS-CoV-2. Molecules 2022; 27: 5405.
- Hsu RL, Lee KT, Wang JH, et al. Amyloid-degrading ability of nattokinase from Bacillus subtilis natto. Journal of Agricultural and Food Chemistry 2009; 57(2): 503-508.
- Ni A, Li H, Wang R, et al. Degradation of amyloid β-peptides catalyzed by nattokinase in vivo and in vitro. Food Science and Human Wellness 2023; 12: 1905-1916.
- Naik S, Katariya R, Shelke S, et al. Nattokinase prevents beta-amyloid peptide (Abeta(1-42)) induced neuropsychiatric complications, neuroinflammation and BDNF signalling disruption in mice. European Journal of Pharmacology 2023; 952: 175821.
- Ladner-Keay CL, Griffith BJ, Wishart DS. Shaking alone induces de novo conversion of recombinant prion proteins to beta-sheet rich oligomers and fibrils. PLoS One 2014; 9(6): e98753.
- Krzek M, Stroobants S, Gelin P, et al. Influence of Centrifugation and Shaking on the Self-Assembly of Lysozyme Fibrils. Biomolecules 2022; 12(12): 1746.
- Hill EK, Krebs B, Goodall DG, et al. Shear flow induces amyloid fibril formation. Biomacromolecules 2006; 7(1): 10-13.
- Dunstan DE, Hamilton-Brown P, Asimakis P, et al. Shear flow promotes amyloid-beta fibrilization. Protein Engineering, Design and Selection 2009; 22(12): 741-746.
- Chaari A, Fahy C, Chevillot-Biraud A, et al. Insights into Kinetics of Agitation-Induced Aggregation of Hen Lysozyme under Heat and Acidic Conditions from Various Spectroscopic Methods. PLoS One 2015; 10(11): e0142095.
- Teoh CL, Bekard IB, Asimakis P, et al. Shear flow induced changes in apolipoprotein C-II conformation and amyloid fibril formation. Biochemistry 2011; 50(19): 4046-4057.
- Dunstan IBBaDE. The Effect of Shear Flow on Amyloid Fibril Formation and Morphology. In Bio-nanoimaging: Protein Misfolding & Aggregation; Elsevier: Amsterdam, The Netherlands, 2014; pp. 503-513.
- Gospodarczyk W, Kozak M. The severe impact of in vivo-like microfluidic flow and the influence of gemini surfactants on amyloid aggregation of hen egg white lysozyme. RSC Advances 2017; 7: 10973.
- Mangione PP, Esposito G, Relini A, et al. Structure, folding dynamics, and amyloidogenesis of D76N beta2-microglobulin: roles of shear flow, hydrophobic surfaces, and alpha-crystallin. Journal of Biological Chemistry 2013; 288(43): 30917-30930.
- Dobson J, Kumar A, Willis LF, et al. Inducing protein aggregation by extensional flow. Proceedings of the National Academy of Sciences of the United States of America 2017; 114(18): 4673-4678.
- Herrera-Rodríguez AM, Dasanna AK, Daday C, et al. The role of flow in the self-assembly of dragline spider silk proteins. Biophysical Journal 2023; 122(21): 4241-4253.
- Almohammadi H, Bagnani M, Mezzenga R. Flow-induced order-order transitions in amyloid fibril liquid crystalline tactoids. Nature Communications 2020; 11(1): 5416.
- Majka Z, Kwiecien K, Kaczor A. Vibrational Optical Activity of amyloid fibrils. ChemPlusChem 2024; 89(8): e202400091.
- Kleiner-Grote GRM, Risse JM, Friehs K. Secretion of recombinant proteins from E. coli. Engineering in Life Sciences 2018; 18(8): 532-550.
- Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990; 96(1): 23-28.
- Schimek C, Egger E, Tauer C, et al. Extraction of recombinant periplasmic proteins under industrially relevant process conditions: Selectivity and yield strongly depend on protein titer and methodology. Biotechnology Progress 2020; 36(5): e2999.
- Mital S, Christie G, Dikicioglu D. Recombinant expression of insoluble enzymes in Escherichia coli: a systematic review of experimental design and its manufacturing implications. Microbial Cell Factories 2021; 20(1): 208.
- Mahler HC, Friess W, Grauschopf U, et al. Protein aggregation: pathways, induction factors and analysis. Journal of Pharmaceutical Sciences 2009; 98(9): 2909-2934.
- Badiei N, Curtis DJ, Campbell AI, et al. Effects of shear flow on the microstructure and elasticity of incipient clots in whole blood and fibrin-thrombin gels. Journal of Thrombosis and Haemostasis 2013; 11: 698-698.
- Badiei N, Sowedan AM, Curtis DJ, et al. Effects of unidirectional flow shear stresses on the formation, fractal microstructure and rigidity of incipient whole blood clots and fibrin gels. Clinical Hemorheology and Microcirculation 2015; 60(4): 451-464.
- Onasoga-Jarvis AA, Puls TJ, O'Brien SK, et al. Thrombin generation and fibrin formation under flow on biomimetic tissue factor-rich surfaces. Journal of Thrombosis and Haemostasis 2014; 12(3): 373-382.
- Bekard IB, Dunstan DE. The Effect of Shear Flow on Amyloid Fibril Formation and Morphology. In Bionanoimaging; Academic Press: New York, NY, USA, 2014; pp. 503-513.
- Eyisoylu H, Hazekamp ED, Cruts J, et al. Flow affects the structural and mechanical properties of the fibrin network in plasma clots. Journal of Materials Science: Materials in Medicine 2024; 35(1): 8.
- Hammarström P, Simon R, Nystrom S, et al. A fluorescent pentameric thiophene derivative detects in vitro-formed prefibrillar protein aggregates. Biochemistry 2010; 49(32): 6838-6845.
- Klingstedt T, Nilsson KPR. Luminescent conjugated poly- and oligo-thiophenes: optical ligands for spectral assignment of a plethora of protein aggregates. Biochemical Society Transactions 2012; 40(4): 704-710.
- König C, Skånberg R, Hotz I, et al. Binding sites for luminescent amyloid biomarkers from non-biased molecular dynamics simulations. Chemical Communications 2018; 54(24): 3030-3033.
- Nilsson KP, Lindgren M, Hammarström P. A pentameric luminescent-conjugated oligothiophene for optical imaging of in vitro-formed amyloid fibrils and protein aggregates in tissue sections. Methods in Molecular Biology 2012; 849: 425-434.
- Nyström S, Bäck M, Nilsson KPR, et al. Imaging Amyloid Tissues Stained with Luminescent Conjugated Oligothiophenes by Hyperspectral Confocal Microscopy and Fluorescence Lifetime Imaging. Journal of Visualized Experiments (JoVE) 2017; 128: 56279.
- Sjölander D, Bijzet J, Hazenberg BPC, et al. Sensitive and rapid assessment of amyloid by oligothiophene fluorescence in subcutaneous fat tissue. Amyloid 2015; 22(1): 19-25.
- Petrlova J, Samsudin F, Bond PJ, et al. SARS-CoV-2 spike protein aggregation is triggered by bacterial lipopolysaccharide. FEBS Letters 2022; 596(19): 2566-2575.
- Morten MJ, Sirvio L, Rupawala H, et al. Quantitative super-resolution imaging of pathological aggregates reveals distinct toxicity profiles in different synucleinopathies. Proceedings of the National Academy of Sciences of the United States of America 2022; 119(41): e2205591119.
- Ahn HJ, Zamolodchikov D, Cortes-Canteli M, et al. Alzheimer's disease peptide beta-amyloid interacts with fibrinogen and induces its oligomerization. Proceedings of the National Academy of Sciences of the United States of America 2010; 107(50): 21812-21817.
- Biza KV, Nastou KC, Tsiolaki PL, et al. The amyloid interactome: Exploring protein aggregation. PLoS One 2017; 12(3): e0173163.
- Kell DB, Pretorius E. Proteomic evidence for amyloidogenic cross-seeding in fibrinaloid microclots. bioRxiv 2024.
- Zamolodchikov D, Berk-Rauch HE, Oren DA, et al. Biochemical and structural analysis of the interaction between beta-amyloid and fibrinogen. Blood 2016; 128(8): 1144-1151.
- Singh PK, Kawasaki M, Berk-Rauch HE, et al. Aminopyrimidine Class Aggregation Inhibitor Effectively Blocks Abeta-Fibrinogen Interaction and Abeta-Induced Contact System Activation. Biochemistry 2018; 57(8): 1399-1409.
- Singh PK, Simoes-Pires EN, Chen ZL, et al. Lecanemab blocks the effects of the Abeta/fibrinogen complex on blood clots and synapse toxicity in organotypic culture. Proceedings of the National Academy of Sciences of the United States of America 2024; 121(17): e2314450121.
- Ma LY, Song JH, Gao PY, et al. Amyloid pathology mediates the associations between plasma fibrinogen and cognition in non-demented adults. Journal of Neurochemistry 2024; 168(9): 2532-2542.
- Benson MD, Liepnieks J, Uemichi T, et al. Hereditary renal amyloidosis associated with a mutant fibrinogen alpha-chain. Nature Genetics 1993; 3(3): 252-255.
- Gillmore JD, Lachmann HJ, Rowczenio D, et al. Diagnosis, pathogenesis, treatment, and prognosis of hereditary fibrinogen A alpha-chain amyloidosis. Journal of the American Society of Nephrology 2009; 20(2): 444-451.
- Stangou AJ, Banner NR, Hendry BM, et al. Hereditary fibrinogen A alpha-chain amyloidosis: phenotypic characterization of a systemic disease and the role of liver transplantation. Blood 2010; 115(15): 2998-3007.
- Haidinger M, Werzowa J, Kain R, et al. Hereditary amyloidosis caused by R554L fibrinogen Aalpha-chain mutation in a Spanish family and review of the literature. Amyloid 2013; 20(2): 72-79.
- Sivalingam V, Patel BK. Familial mutations in fibrinogen Aalpha (FGA) chain identified in renal amyloidosis increase in vitro amyloidogenicity of FGA fragment. Biochimie 2016; 127: 44-49.
- Yazaki M, Yoshinaga T, Sekijima Y, et al. Hereditary Fibrinogen Aalpha-Chain Amyloidosis in Asia: Clinical and Molecular Characteristics. International Journal of Molecular Sciences 2018; 19(1): 320.
- Chapman J, Dogan A. Fibrinogen alpha amyloidosis: insights from proteomics. Expert Review of Proteomics 2019; 16(9): 783-793.
- Jin S, Shen Z, Li J, et al. Fibrinogen A Alpha-Chain Amyloidosis Associated With a Novel Variant in a Chinese Family. Kidney International Reports 2021; 6(10): 2726-2730.
- Li ZY, Wang S, Li DY, et al. Fibrinogen A Alpha-Chain Amyloidosis in Two Chinese Patients. Frontiers in Medicine 2022; 9: 869409.
- Eriksson M, Schönland S, Bergner R, et al. Three German fibrinogen Aalpha-chain amyloidosis patients with the p.Glu526Val mutation. Virchows Archiv 2008; 453(1): 25-31.
- de Villiers S, Bester J, Kell DB, et al. Erythrocyte health and the possible role of amyloidogenic blood clotting in the evolving haemodynamics of female migraine-with-aura pathophysiology: Results from a pilot study. Frontiers in Neurology 2019; 10: 1262.
- Kruger A, Vlok M, Turner S, et al. Proteomics of fibrin amyloid microclots in Long COVID/Post-Acute Sequelae of COVID-19 (PASC) shows many entrapped pro-inflammatory molecules that may also contribute to a failed fibrinolytic system. Cardiovascular Diabetology 2022; 21: 190.
- Page MJ, Thomson GJA, Nunes JM, et al. Serum amyloid A binds to fibrin(ogen), promoting fibrin amyloid formation. Scientific Reports 2019; 9(1): 3102.
- Pretorius E, Venter C, Laubscher GJ, et al. Prevalence of readily detected amyloid blood clots in ‘unclotted’ Type 2 Diabetes Mellitus and COVID-19 plasma: A preliminary report. Cardiovascular Diabetology 2020; 19: 193.
- Turner S, Laubscher GJ, Khan MA, et al. Rapid flow cytometric analysis of fibrin amyloid microclots in Long COVID. Research Square 2023; preprint; doi:10.21203/rs.3.rs-2731434/v1.
- Visser MJE, Pretorius E. Atomic Force Microscopy: The Characterisation of Amyloid Protein Structure in Pathology. Current Topics in Medicinal Chemistry 2019; 19(32): 2958-2973.
- Pretorius E, Nunes M, Pretorius J, et al. Flow Clotometry: Measuring Amyloid Microclots in ME/CFS, Long COVID, and Healthy Samples with Imaging Flow Cytometry. Research Square 2024; preprint; doi:10.21203/rs.3.rs-4507472/v1.
- Meisl G, Knowles TP, Klenerman D. The molecular processes underpinning prion-like spreading and seed amplification in protein aggregation. Current Opinion in Neurobiology 2020; 61: 58-64.
- Young KA, Mancera RL. Review: Investigating the aggregation of amyloid beta with surface plasmon resonance: Do different approaches yield different results? Analytical Biochemistry 2022; 654: 114828.
- Jan A, Gokce O, Luthi-Carter R, et al. The ratio of monomeric to aggregated forms of Abeta40 and Abeta42 is an important determinant of amyloid-beta aggregation, fibrillogenesis, and toxicity. Journal of Biological Chemistry 2008; 283(42): 28176-28189.
- Wang L, Eom K, Kwon T. Different Aggregation Pathways and Structures for Abeta40 and Abeta42 Peptides. Biomolecules 2021; 11(2): 198.
- Wei G, Su Z, Reynolds NP, et al. Self-assembling peptide and protein amyloids: from structure to tailored function in nanotechnology. Chemical Society Reviews 2017; 46(15): 4661-4708.
- Tipping KW, van Oosten-Hawle P, Hewitt EW, et al. Amyloid Fibres: Inert End-Stage Aggregates or Key Players in Disease? Trends in Biochemical Sciences 2015; 40(12): 719-727.
- Tycko R. Structure of aggregates revealed. Nature 2016; 537(7621): 492-493.
- Bunce SJ, Wang Y, Stewart KL, et al. Molecular insights into the surface-catalyzed secondary nucleation of amyloid-beta(40) (Abeta(40)) by the peptide fragment Abeta(16-22). Science Advances 2019; 5(6): eaav8216.
- Michaels TCT, Saric A, Habchi J, et al. Chemical Kinetics for Bridging Molecular Mechanisms and Macroscopic Measurements of Amyloid Fibril Formation. Annual Review of Physical Chemistry 2018; 69: 273-298.
- Vaquer-Alicea J, Diamond MI. Propagation of Protein Aggregation in Neurodegenerative Diseases. Annual Review of Biochemistry 2019; 88: 785-810.
- Ke PC, Zhou R, Serpell LC, et al. Half a century of amyloids: past, present and future. Chemical Society Reviews 2020; 49(15): 5473-5509.
- Riedel CH, Zimmermann P, Jensen-Kondering U, et al. The importance of size: successful recanalization by intravenous thrombolysis in acute anterior stroke depends on thrombus length. Stroke 2011; 42(6): 1775-1777.
- Kamalian S, Morais LT, Pomerantz SR, et al. Clot length distribution and predictors in anterior circulation stroke: implications for intra-arterial therapy. Stroke 2013; 44(12): 3553-3556.
- Ganeshan R, Nave AH, Scheitz JF, et al. Assessment of thrombus length in acute ischemic stroke by post-contrast magnetic resonance angiography. Journal of NeuroInterventional Surgery 2018; 10(8): 756-760.
- Rossi R, Fitzgerald S, Gil SM, et al. Correlation between acute ischaemic stroke clot length before mechanical thrombectomy and extracted clot area: Impact of thrombus size on number of passes for clot removal and final recanalization. European Stroke Journal 2021; 6(3): 254-261.
- Patki P, Simon S, Manning KB, et al. Computational analysis of effects of clot length on Acute ischemic stroke recanalization under different cyclic aspiration loading conditions. International Journal for Numerical Methods in Biomedical Engineering 2023; 39(2): e3667.
- Currin A, Swainston N, Day PJ, et al. Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently. Chemical Society Reviews 2015; 44(5): 1172-1239.
- Yang HR, Zahan MN, Yoon Y, et al. Unveiling the Potent Fibrino(geno)lytic, Anticoagulant, and Antithrombotic Effects of Papain, a Cysteine Protease from Carica papaya Latex Using kappa-Carrageenan Rat Tail Thrombosis Model. International Journal of Molecular Sciences 2023; 24(23): 16770.
- Metkar SK, Girigoswami A, Murugesan R, et al. Lumbrokinase for degradation and reduction of amyloid fibrils associated with amyloidosis. Journal of Applied Biomedicine 2017; 15(2): 96-104.
- Metkar SK, Ghosh S, Girigoswami A, et al. The Potential of Serratiopetidase and Lumbrokinase for the Degradation of Prion Peptide 106-126—an In Vitro and In Silico Perspective. CNS & Neurological Disorders - Drug Targets 2019; 18(9): 723-731.
- Metkar SK, Girigoswami A, Vijayashree R, et al. Attenuation of subcutaneous insulin induced amyloid mass in vivo using Lumbrokinase and Serratiopeptidase. International Journal of Biological Macromolecules 2020; 163: 128-134.
- Fadl NN, Ahmed HH, Booles HF, et al. Serrapeptase and nattokinase intervention for relieving Alzheimer's disease pathophysiology in rat model. Human & Experimental Toxicology 2013; 32(7): 721-735.
- Jadhav SB, Shah N, Rathi A, et al. Serratiopeptidase: Insights into the therapeutic applications. Biotechnology Reports (Amsterdam, Netherlands) 2020; 28: e00544.
- Metkar SK, Girigoswami A, Bondage DD, et al. The potential of lumbrokinase and serratiopeptidase for the degradation of Aβ 1–42 peptide—an in vitro and in silico approach. International Journal of Neuroscience 2024; 134: 112-123.
- Liao J, Ren X, Yang B, et al. Targeted thrombolysis by using c-RGD-modified N,N,N-Trimethyl Chitosan nanoparticles loaded with lumbrokinase. Drug Development and Industrial Pharmacy 2019; 45(1): 88-95.
- Metkar SK, Girigoswami A, Murugesan R, et al. In vitro and in vivo insulin amyloid degradation mediated by Serratiopeptidase. Materials Science & Engineering C - Materials for Biological Applications 2017; 70(Pt 1): 728-735.
- Nair SR, C SD. Serratiopeptidase: An integrated View of Multifaceted Therapeutic Enzyme. Biomolecules 2022; 12(10): 1468.
- Katsipis G, Avgoulas DI, Geromichalos GD, et al. In vitro and in silico evaluation of the serrapeptase effect on biofilm and amyloids of Pseudomonas aeruginosa. Applied Microbiology and Biotechnology 2023; 107(23): 7269-7285.
- Mei Jf, Cai Sf, Yi Y, et al. Study of the fibrinolytic activity of serrapeptase and its in vitro thrombolytic effects. Brazilian Journal of Pharmaceutical Sciences 2022; 58: e201004.
- Banerjee G, Ray AK. Impact of microbial proteases on biotechnological industries. Biotechnology & Genetic Engineering Reviews 2017; 33(2): 119-143.