Main Text
1 Introduction
Cancer is a problem for public health worldwide. According to statistics, there were 1,670 cancer-related fatalities and 5,370 new cases of cancer identified in America in 2023. Nevertheless, over the previous ten years, the death rate from CRC has declined annually by 2% [1]. CRC is a malignant tumor occurring from the cecum to the rectum, including rectal cancer and colon cancer, which is one of the three most common cancers globally. According to the 2020 global cancer statistics, CRC had the third highest incidence [2] and was the second leading cause of death for women [3,4]. The main causes of CRC are long-term inflammatory bowel illness, personal and family medical histories, prolonged inactivity, poor diet, and alcohol consumption [5]. Nowadays, surgical resection combined with chemotherapy is the mainstay of CRC treatment. However, the 5-year survival rate is low, and chemotherapy has significant hazardous side effects [6]. Hence, new treatment schemes and strategies are of great significance, and notably, TCM combined with chemotherapy has unique advantages in the prognosis of CRC patients [7,8].
TCM offers a broad range of opportunities for disease prevention and therapy with its emphasis on holism and extensive treatment history with various ingredients [9]. Many studies have been conducted on anti-CRC properties of TCM and how important they are for treating and preventing CRC. TCM was reported that it inhibits angiogenesis and CRC cell proliferation, boosts immunity, and prolongs CRC patient lifespan [10]. The capacity of TCM to regulate a variety of signaling pathways, such as Phosphatidylinositol 3-kinase/protein kinase-B (PI3K/Akt) signaling pathway, Nuclear factor kappa-beta (NF-κB) signaling pathway, Mitogen-activated protein kinase (MAPK) signaling pathway, and immunomodulatory signaling pathways. Such regulation impacts key biological processes like cell proliferation, apoptosis, cell cycle progression, and chemoresistance, thereby exerting a multifaceted therapeutic influence on CRC [10]. Due to the complex mechanism of TCM in CRC patients, its active ingredients and action mechanisms need systematic exploration. In recent years, with the combination of new technologies and TCM research, achievements have been made in research on the modernization of TCM. Network medicine is an emerging discipline based on computer technology, systems biology network analysis, and multi-target drug molecular design [11], which features “holism” and “systematization” [12,13]. This technology provides new possibilities for unveiling the multiple mechanisms of TCM in treating CRC.
This study (Figure 1) summarized the literature about anti-CRC TCM in the PubMed database and identified the potential TCM that can effectively treat CRC. Later, the active ingredients and mode of action of TCM in the treatment of CRC were identified by network medical analysis, offering a theoretical foundation and empirical support for the use of TCM in the treatment of CRC.

Figure 1 Detail chart of this study.
2 Methods
2.1 Collection of anti-CRC TCM
To obtain effective anti-CRC TCM, literature about the treatment of CRC with TCM was collected through text analysis on the PubMed database. A total of 14,558 pieces of literature on CRC were retrieved with "Colorectal cancer", "Colorectal neoplasms", "Colorectal tumors", "Colorectal carcinoma" and "Colorectal" as keywords, and by limiting keywords in the title/abstract. Regarding the previously published report, "Herbal medicine" and "Colorectal cancer" were used as keywords [14], and 83 TCMs that were reported to be applied to treat CRC were retrieved (Data deadline: September 2023). Next, the correlation between TCM and CRC was measured according to a reported formula [15], using R language software version 4.3.0 to rank the correlation of TCM. The formula was as follows, where N represents the total number of articles on the PubMed database, n represents the number of articles involving a single TCM, K represents the total number of articles involving CRC, and k represents the number of articles involving a single TCM related to CRC.
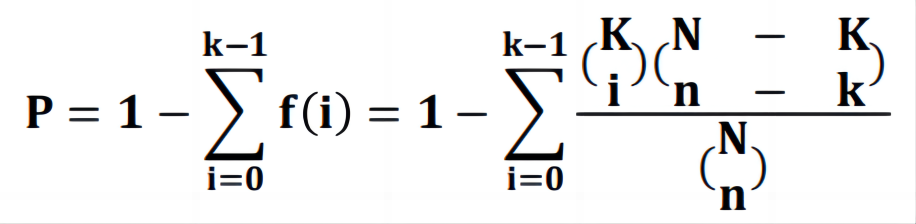
2.2 Screening of active ingredients in TCM
The ingredients of TCM were acquired using the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, https:// www.tcmsp-e.com/) and HERB databases (a high-throughput experiment- and reference-guided database of traditional Chinese medicine, http://herb.ac.cn) [16,17]. Of them, TCMSP combined with kinetic parameters was applied to set oral bioavailability (OB) ≥ 30 and drug-likeness (DL) ≥ 0.18, to screen out qualified TCM ingredients. Then, these ingredients were summarized, duplicates were removed, and the data was saved in Excel for subsequent data processing.
2.3 Screening of targets active ingredients of indicated TCM
In this paper, the collected ingredients were analyzed on DrugBank (https://go.drugbank.com/), TTD (http://db.idrblab.net/ttd/), BindingDB (https://www.bindingdb.org/bind/index.jsp), and IPA software to obtain relevant component targets. The ingredient targets from each database were summarized to remove duplicates and were converted into gene names using the UniProt database (https://www.uniprot.org/). The species was limited to "Homo sapiens". The obtained gene names were converted into Gene ID via NCBI database (https://www.ncbi.nlm.nih.gov/) for further calculation of network proximity. Detailed ingredients and target information are shown in Supplementary Material 1.
2.4 Collection of CRC genes
Using "Colorectal cancer" as the keyword, genes associated with CRC were retrieved through 4 databases CTD (http://ctdbase.org), TTD, DrugBank, and PharmGKB (http://www.pharmgkb.org). The ingredient targets and CRC-related genes were imported into the online tool Venny 2.1.0 (http://bioinformatics.psb.ugent.be/), and the intersection part was the potential target of TCM in treating CRC. Detailed disease genes were shown in Supplementary Material 2.
2.5 Calculation of network proximity
Network proximity is the shortest distance (dc) between drug targets (T) and disease-related genes (S). In line with a former study, the dc between T and S in the human interaction group was calculated, and then a reference distribution distance was established to evaluate the importance of drugs and diseases. T and S with the same network distance were calculated 1000 times to obtain the Z-Score [18]. The online calculation website is http://www.zmupredict.cn/proximity. When the Z-Score < -1.5 and p ≤ 0.05, the TCM ingredients were considered to have a pharmacologic action. For more details, please refer to our previous research [19,20].
2.6 Protein-protein interaction (PPI) network diagram
The intersecting targets of TCM ingredients and CRC were collected through Venny 2.1.0, and the potential targets were imported into String database (https://www.string-db.org/), with the species limited to "Homo Sapiens". Edge and node data were saved and imported into Gephi 0.9.2 software to plot PPI network diagram.
2.7 Plotting of TCM-ingredients-targets diagram
The data were imported into Gephi 0.9.2 software to create a visualized TCM-ingredients-targets diagram.
2.8 GO enrichment and KEGG pathway analyses of anti-CRC TCMs
To acquire GO enrichment and KEGG pathway analyses of TCM ingredients targets and disease-related genes, the intersecting targets of TCM and disease were imported into DAVID database (https://david.ncifcrf.gov/), with the species limited to "Homo Sapiens". Then, the main pathways and annotated genes were singled out, and microbioinformatics (a plotting website) was employed to conduct visualized analyses. GO enrichment covers three types, namely molecular function (MF), biological process (BP), and cellular component (CC).
3 Results and analyses
3.1 The top ten TCMs highly correlated with CRC
Through literature collection, correlation calculation, and ranking, the top ten TCMs highly correlated with CRC were identified, including Panax quinquefolium L., Scutellaria barbata D.Don, Panax Ginseng C. A. Mey., Salvia miltiorrhiza Bge., Ganoderma lucidum, Zingiber officinale Rosc., Salvia japonica Thunb., Glycyrrhiza uralensis Fisch, Sophora flavescens Ait and Paris polyphylla Smith var.chinensis (Franch.) Hara (Table 1). Among them, Panax quinquefolium L. ranked first, accounting for 38.60% (p = 7.51E-60) and Scutellaria barbata D.Don ranked second, accounting for 11.31% (p = 1.70E-40). p ≤ 0.01 represents a high correlation. Therefore, these TCMs had a remarkable anti-CRC effect.
Table 1 The top ten TCMs highly correlated with CRC treatment.
| Latin name | Chinese name | Total | Relevant to CRC | Rate (%) | p-value |
|---|---|---|---|---|---|
| Panax quinquefolium L. | Xiyangshen | 57 | 22 | 38.60 | 7.51E-60 |
| Scutellaria barbata D.Don | Banzhilian | 168 | 19 | 11.31 | 1.70E-40 |
| Panax Ginseng C. A. Mey. | Renshen | 5425 | 31 | 0.57 | 4.58E-25 |
| Salvia miltiorrhiza Bge. | Danshen | 136 | 12 | 8.82 | 9.14E-25 |
| Ganoderma lucidum | Lingzhi | 2461 | 23 | 0.93 | 1.14E-23 |
| Zingiber officinale Rosc. | Shengjiang | 751 | 14 | 1.86 | 4.23E-19 |
| Salvia japonica Thunb | Gouweicao | 172 | 10 | 5.81 | 5.20E-19 |
| Glycyrrhiza uralensis Fisch | Gancao | 1102 | 13 | 1.18 | 2.61E-15 |
| Sophora flavescens Ait | Kushen | 312 | 9 | 2.88 | 1.75E-14 |
| Paris polyphylla Smith var.chinensis(Franch.)Hara | Chonglou | 270 | 8 | 2.96 | 4.04E-13 |
3.2 Panax quinquefolium L.-target network diagram
TCM features multi-ingredients and multi-targets, which commonly perform extensive pharmacologic actions through multiple ingredients and targets. Panax quinquefolium L. having the highest correlation with CRC, was selected for subsequent network visualized analyses. According to Figure 2, there were 202 nodes and 255 edges. A greater number of ingredient targets mean a higher degree. Figure 2 only displayed labels with nodes of degree ≥ 2.

Figure 2 TCM-ingredients-targets network diagram. (Red: Panax quinquefolium L.; Blue: TCM ingredients; Pink: TCM targets.)
3.3 Calculation results regarding the network proximity of TCM-targets and CRC-related genes
Concerning an existing report, we calculated the shortest distance between TCM ingredient targets and CRC-related genes in the human interaction group to evaluate the therapeutic efficacy of active ingredients on CRC. The ingredients and targets of Panax quinquefolium L. were collected from a public database. There were 39 ingredients with 162 targets, and 186 CRC-related genes. The results were exhibited in Table 2. The ranking was completed based on Z-Score. A lower Z-Score signifies a closer distance and a high correlation. Our results indicated that Z-Scores of Ginsenoside Rb3, Papaverine, Ginsenoside Rg3, and Ginsenoside Rb1 were all below -1.5, which may be the key ingredients of Panax quinquefolium L. for anti-CRC. Detailed results of the network proximity calculations were presented in Supplementary Material 3.
Table 2 The predicted top ten active components of Panax quinquefolium L. based on network proximity.
| Ingredient | Chemical structure | Z-score | p-value | Related literature |
|---|---|---|---|---|
| Ginsenoside rb3 | 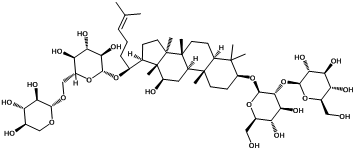 | -4.29 | <0.001 | [19] |
| Papaverine | 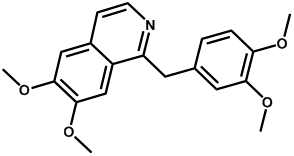 | -4.16 | <0.001 | - |
| Ginsenoside rg3 | 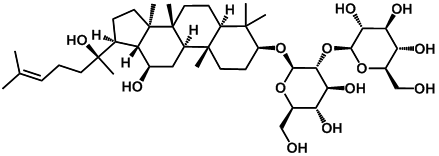 | -4.12 | <0.001 | [20] |
| Ginsenoside rb1 | 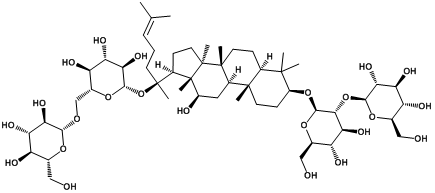 | -3.38 | <0.001 | [21] |
| Oleanolic acid | 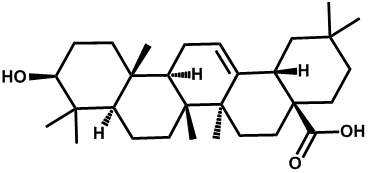 | -3.35 | <0.001 | [22] |
| Hexadecanoic acid | 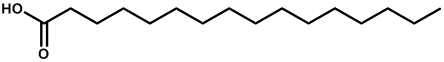 | -3.31 | 0.003 | [23] |
| Ginsenoside rd | 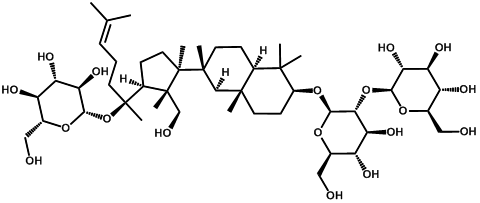 | -3.15 | <0.001 | [24] |
| Linolenic acid | 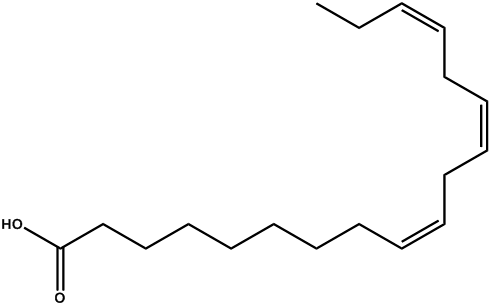 | -3.08 | 0.004 | [25] |
| Capsaicin | 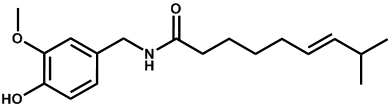 | -3.01 | 0.001 | [26] |
| Lecithin | 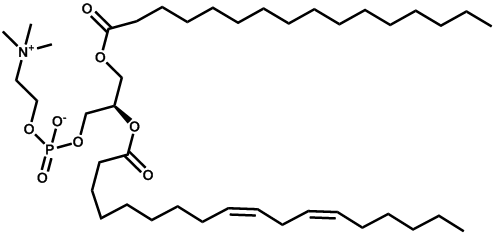 | -1.95 | 0.019 | - |
"-" means no directly relevant reports.
3.4 Venn diagram of "TCM-disease" targets and PPI analyses
Biological functions or pathways may play roles in the regulation of relevant proteins. We adopted PPI to elucidate the effective ingredients and mechanism of Panax quinquefolium L. for anti-CRC. 12 intersecting targets were obtained after the retrieved 162 ingredient targets and 186 CRC-related genes were made into Venn diagram (Figure 3). Then, the intersecting targets were imported into String database to obtain the interaction diagram of potential targets. The results revealed that epidermal growth factor receptor (EGFR) (degree = 7), protein kinase C alpha (PRKCA) (degree = 6), and prostaglandin-endoperoxide synthase 2 (PTGS2) (degree = 6) had relatively high degrees, which may be the critical genes to treat CRC.

Figure 3 Venn diagram of Panax quinquefolium L. against CRC and PPI analyses. (A) Intersecting targets of component targets and CRC-related genes; (B) PPI network diagram.
3.5 GO enrichment and KEGG pathway analysis results
To thoroughly explore the biological function of Panax quinquefolium L. in treating CRC, we used DAVID online database to conduct GO functional enrichment and KEGG pathway analyses, and created GO and KEGG bubble diagrams using microbioinformatics online plotting website. The top five BP, CC, and MF were subjected to GO enrichment analyses based on Count value (Figure 4). These functions predominantly encompass the positive regulation of transcription from RNA polymerase II promoter, cytoplasm, and protein binding. Furthermore, we selected the top ten pathways for KEGG pathway analysis based on their count values. The resulting bubble diagram, presented in Figure 5, highlights the gene enrichment pathways relevant to CRC treatment. These pathways predominantly involve mechanisms in cancer, microRNAs (miRNAs) in cancer, human cytomegalovirus infection, and the Rap1 signaling pathway.

Figure 4 GO enrichment analysis of potential targets. (Circle: BP; Triangle: CC: Quadrate: MF.)

Figure 5 Bubble diagram of KEGG pathway analysis. Larger bubble and darker color imply stronger correlation of the pathway.
4 Conclusion and perspective
This study examined the active ingredients and mechanisms based on the top-ranked TCM, as well as the promising anti-CRC TCMs, using text mining and network medicine. Ten TCMs, including Panax quinquefolium L., Scutellaria barbata D.Don, and Panax ginseng C. A. Mey, showed a strong connection with CRC, according to text mining results. Panax quinquefolium L. can tonify qi, nourish yin, clear heat, and produce fluid; Scutellaria barbata D.Don can clear heat, remove toxins, resolve stasis, and promote urination; and Panax Ginseng C. A. Mey can tonify the spleen and replenish the lung, according to the efficacy of TCMs documented in the Chinese Pharmacopoeia (2020 edition). TCM emphasizes "treatment with both elimination and reinforcement" in accordance with the principle of "treatment based on syndrome differentiation". In addition to suppressing cancer cell growth, TCM also improves physical health and immunity to help intestinal cancer patients recover better and faster. The subsequent analyses were performed on TCM which had the highest correlation with CRC. The results of network proximity showed that Ginsenoside Rb3, Papaverine, Ginsenoside Rg3, and Ginsenoside Rb1 may be the key active ingredients to treat CRC, and EGFR, PRKCA, and PTGS2 were potential targets. KEGG pathway analysis data indicated that pathways in cancer are pivotal anti-CRC pathways. These findings theoretically guide the follow-up clinical medication and provide novel research thoughts to expound on the active ingredients and mechanisms of TCM in treating CRC.
In previous reports, Panax quinquefolium L. and its extract have been widely applied to treat cardiovascular diseases [27], diabetes, and chronic obstructive pulmonary disease [28,29]. The main ingredients of Panax quinquefolium L. are Ginsenosides (Rb1, Rb2, Rg3, Re, and Rd), and consistent with previous reports, we found that Ginsenoside Rb3, Ginsenoside Rg3, Ginsenoside Rb1, and Ginsenoside Rd were ranked in the top ten of active ingredients in Panax quinquefolium L. Ginsenoside Rg3 can be used to treat various CRC cells, which has a strong therapeutic effect through mediating protein expressions of C/EBPβ/NF-κB pathway to dampen CRC cell proliferation [21,22]. Relative to other saponins, Ginsenoside Rb3 has a weak effect on anti-CRC, 300 μM of which barely affecting CRC cell proliferation [21]; however, by anti-inflammation and anti-oxidant stress effects, Ginsenoside Rb3 can impact other diseases, such as exerting protective properties against cigarette smoke extract-induced cell injury and fibrosis [30]. Moreover, Ginsenoside Rb3 has been confirmed to play an anti-cancer role by restoring mucosal structure, improving mucosal immunity, promoting probiotic growth, and reducing relevant flora [31]. It has been documented that Ginsenoside Rb1 can inhibit the proliferation of SW480 cells [32], which similarly decreases myocardial infarction area while attenuating fibrosis and reperfusion injury through inhibiting reactive oxygen species production from mitochondrial complex I [33]. Ginsenoside Rd has been verified to inhibit CRC cell migration via EGFR [26]. Furthermore, in the calculation of our top ten ingredients, fatty acids were also included, such as linolenic acid, hexadecanoic acid, and oleanolic acid. Reportedly, these ingredients can suppress tumor cell growth based on different mechanisms. Linolenic acid plays an anti-cancer role through declining oxidative phosphorylation and glycolysis to render the tumor cells in a resting state [27]. Oleanolic acid is capable of dampening the proliferation of tumors in vivo and in vitro [24]. Hexadecanoic acid can treat cancer by blocking epithelial-mesenchymal transition in CRC cells and reducing tumor-related macrophages [25]. Besides, papaverine occupied the second position in active ingredients, with multiple pharmacologic actions, such as anti-cancer, anti-inflammation, and neuroprotection. The papaverine derivative caroverine inhibits the expression of VEGF in CRC cells to attenuate the toxic effect of linoleic acid hydroperoxide on cells [32]. Pepper is composed of about 69% capsaicin which has anti-inflammation and anti-oxidant properties, and can treat CRC by mediating cell cycle arrest, proliferation, and apoptosis [28]. Lecithin is mainly derived from plant species, including soybeans, sunflowers, and canola seed, which has functions in micelles and oil-in-water emulsions and can improve the rate of drug release and administration mode [34,35], but little is reported about its therapeutic effect on cancer. Collectively, the first nine of the top ten ingredients have anti-cancer effects, and thus this method has certain advantages in predicting drugs.
EGFR is a kind of transmembrane protein, belonging to the Erythroblastic Leukemia Viral Oncogene Homolog (ERBB) family, the expression level of which in CRC patients has an intimate association with tumor cell proliferation. In in-vitro CRC cell models, knocking down EGFR expression can inhibit CRC cell proliferation and migration [36]. Therefore, EGFR has been extensively considered to be the promising target of CRC treatment. Additionally, PRKCA suppresses WNT activity to increase the survival rate of CRC patients [37]. PTGS2 is not expressed on normal rectal mucosa, but the deficiency of PTGS2 is more likely to cause neutrophil infiltration or slow ulcer healing, ultimately inducing CRC [38]. There is a study unveiling that PTGS2 is remarkably overexpressed in tumor tissues [39]. In our prediction results, EGFR, PRKCA, and PTGS2 had high degrees, which may be the critical genes to treat CRC.
Next, we analyzed KEGG pathways and GO enrichment for 12 targets. The three primary components of GO enrichment analysis are protein binding, cytoplasm, and positive regulation of transcription from the RNA polymerase II promoter. EGFR tyrosine kinase inhibitor resistance, Rap1 signaling routes, HIF-1 signaling pathways, cancer-related mRNA pathways, and other pathways are the main pathways in KEGG pathway analysis. Various tumor cells ability to proliferate, migrate, and remain viable can be inhibited by miRNA; in fact, people with CRC have much higher levels of miRNA in their plasma. MiRNA may therefore be used as a prospective biomarker for CRC screening and treatment planning [40,41]. According to a former report, Mex3a is remarkably high expressed in tumor tissues and mediates CRC cell proliferation and migration via the Rap1 signaling pathway [42]. Tumor development and metastasis may be inhibited by HIF-1 downregulation [43]. EGFR is the key factor for cell proliferation and differentiation, and despite its prominent role in CRC chemotherapy, the effect of EGFR monoclonal antibody is often limited by drug resistance [44]. It has been shown that tyrosine kinase inhibitors can reduce the resistance of anti-EGFR treatments in colorectal cancer [45]. These pathways have a close relationship with CRC and provide references for subsequent research on the pathogenesis of CRC.
This research utilizes text mining and advanced network medical computing to identify potential anti-colorectal cancer (CRC) Traditional Chinese Medicines (TCMs). It further conducts in-depth network medical computing and visual analysis, focusing specifically on the TCM exhibiting the highest correlation with CRC. A notable limitation of this study is the lack of extensive in vitro or in vivo experimental validation. Consequently, further elucidation of the underlying mechanisms is warranted. TCM, known for its low toxicity, fewer side effects, and multi-ingredient, multi-target characteristics, presents a compelling area of study. Employing network medicine techniques, this study determines the network proximity between active TCM ingredients and illness-related genes, where a lower Z-score signifies greater relevance. This approach offers a valuable framework for assessing the potential therapeutic efficacy of TCM monomers. Ultimately, this research comprehensively delineates the active ingredients, targets, and effective pathways of TCM, thereby contributing significantly to the clinical application and experimental investigation of anti-CRC TCMs.
Back Matter
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
Conceptualization: A.R. and M.L.; Data curation: M.H. and S.Z.; Formal analysis: Y.F. and W.S.; Methodology: A.R. and M.L.; Writing – original draft: M.H. and S.Z.; Writing – review and editing: Y.F. and W.S. All authors have read and agreed to the published version of manuscript.
Ethics Approval and Consent to Participate
The study was approved by the Medical Ethics Committee, and the patients were informed and consented.
Funding
This study is supported by funding from the Traditional Chinese Medicine Bureau of Guangdong Province (Grant NO. 20221330) and the District Health System Research Project of Shenzhen Futian (Grant NO. FTWS2023071).
Availability of Data and Materials
The analyzed data sets generated during the study are available from the corresponding author upon reasonable request.
Supplementary Materials
The following supporting information can be downloaded at: https://ojs.exploverpub.com/index.php/jecacm/article/view/184/sup. Supplementary Material 1: A list of ingredients and targets. Supplementary Material 2: A list of CRC genes. Supplementary Material 3: A list of Network proximity.
References
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA: A Cancer Journal for Clinicians 2023; 73(1): 17-48.
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians 2021; 71(3): 209-249.
- Koch T, Jørgensen JT, Christensen J, et al. Bilateral oophorectomy and rate of colorectal cancer: A prospective cohort study. International Journal of Cancer 2022; 150(1): 38-46.
- Dinami R, Porru M, Amoreo CA, et al. TRF2 and VEGF-A: an unknown relationship with prognostic impact on survival of colorectal cancer patients. Journal of Experimental & Clinical Cancer Research 2020; 39(1): 111.
- Li J, Ma X, Chakravarti D, et al. Genetic and biological hallmarks of colorectal cancer. Genes & Development 2021; 35(11-12): 787-820.
- Qi F, Zhao L, Zhou A, et al. The advantages of using traditional Chinese medicine as an adjunctive therapy in the whole course of cancer treatment instead of only terminal stage of cancer. BioScience Trends 2015; 9(1): 16-34.
- Yang MD, Zhou WJ, Chen XL, et al. Therapeutic Effect and Mechanism of Bushen-Jianpi-Jiedu Decoction Combined with Chemotherapeutic Drugs on Postoperative Colorectal Cancer. Frontiers in Pharmacology 2021; 12: 524663.
- Zheng L, Sha J. Progress in the Application of Traditional Chinese Medicine Treatment in Postoperative Rehabilitation of Colorectal Cancer. Academic Journal of Medicine & Health Sciences 2019; 4(7): 51-57.
- Li S, Wu Z, Le W. Traditional Chinese medicine for dementia. Alzheimer's & Dementia 2021; 17(6): 1066-1071.
- Chen J-F, Wu S-W, Shi Z-M, et al. Traditional Chinese medicine for colorectal cancer treatment: potential targets and mechanisms of action. ChenMed 2023; 18(1): 14-45.
- Zhao L, Zhang H, Li N, et al. Network pharmacology, a promising approach to reveal the pharmacology mechanism of Chinese medicine formula. Journal of Ethnopharmacology 2023; 309: 116306.
- Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nature Chemical Biology 2008; 4(11): 682-690.
- Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chinese Journal of Natural Medicines 2013; 11(2): 110-120.
- Huang C, Zheng C, Li Y, et al. Systems pharmacology in drug discovery and therapeutic insight for herbal medicines. Briefings in Bioinformatics 2014; 15(5): 710-733.
- Fang J, Wang L, Wu T, et al. Network pharmacology-based study on the mechanism of action for herbal medicines in Alzheimer treatment. Journal of Ethnopharmacology 2017; 196: 281-292.
- Fang S, Dong L, Liu L, et al. HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Research 2021; 49(D1): D1197-D1206.
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics 2014; 16: 13.
- Guney E, Menche J, Vidal M, et al. Network-based in silico drug efficacy screening. Nature Communications 2016; 7: 10331.
- Ren A, Ma M, Liang Y, et al., GSZ Formula Enhances Sleep Quality: Exploring Its Active Ingredients and Mechanism Using a Network Medicine Approach. Clinical Complementary Medicine and Pharmacology 2024; 4(1): 100107.
- Ren A, Wu T, Wang Y, et al. Integrating animal experiments, mass spectrometry and network-based approach to reveal the sleep-improving effects of Ziziphi Spinosae Semen and γ-aminobutyric acid mixture. Chinese Medicine 2023; 18(1): 99.
- Wang CZ, Zhang B, Song WX, et al. Steamed American ginseng berry: ginsenoside analyses and anticancer activities. Journal of Agricultural and Food Chemistry 2006; 54(26): 9936-9942.
- Yang X, Zou J, Cai H, et al. Ginsenoside Rg3 inhibits colorectal tumor growth via down-regulation of C/EBPbeta/NF-kappaB signaling. Biomedicine & Pharmacotherapy 2017; 96: 1240-1245.
- Wang CZ, Xie JT, Fishbein A, et al. Antiproliferative effects of different plant parts of Panax notoginseng on SW480 human colorectal cancer cells. Phytotherapy Research 2009; 23(1): 6-13.
- Li LI, Wei L, Shen A, et al. Oleanolic acid modulates multiple intracellular targets to inhibit colorectal cancer growth. International Journal of Oncology 2015; 47(6): 2247-2254.
- De Araujo Junior RF, Eich C, Jorquera C, et al. Ceramide and palmitic acid inhibit macrophage-mediated epithelial–mesenchymal transition in colorectal cancer. Molecular and Cellular Biochemistry 2020; 468(1-2): 153-168.
- Phi LTH, Sari IN, Wijaya YT, et al. Ginsenoside Rd Inhibits the Metastasis of Colorectal Cancer via Epidermal Growth Factor Receptor Signaling Axis. IUBMB Life 2019; 71(5): 601-610.
- Ogata R, Mori S, Kishi S, et al. Linoleic Acid Upregulates Microrna-494 to Induce Quiescence in Colorectal Cancer. International Journal of Molecular Sciences 2021; 23(1): 225.
- Jin J, Lin G, Huang H, et al. Capsaicin Mediates Cell Cycle Arrest and Apoptosis in Human Colon Cancer Cells via Stabilizing and Activating p53. International Journal of Biological Sciences 2014; 10(3): 285-295.
- Sarhene M, Ni JY, Duncan ES, et al. Ginsenosides for cardiovascular diseases; update on pre-clinical and clinical evidence, pharmacological effects and the mechanisms of action. Pharmacological Research 2021, 166: 105481.
- Wang M, Chen X, Jin W, et al. Ginsenoside Rb3 exerts protective properties against cigarette smoke extract-induced cell injury by inhibiting the p38 MAPK/NF-kappaB and TGF-beta1/VEGF pathways in fibroblasts and epithelial cells. Biomedicine & Pharmacotherapy 2018; 108: 1751-1758.
- Huang G, Khan I, Li X, et al. Ginsenosides Rb3 and Rd reduce polyps formation while reinstate the dysbioticgut microbiota and the intestinal microenvironment in ApcMin/+ mice. Scientific Reports 2017; 7(1): 12552.
- Nohl H, Rohr-Udilova N, Gille L, et al. Ubiquinol and the papaverine derivative caroverine prevent the expression of tumour- promoting factors in adenoma and carcinoma colon cancer cells induced by dietary fat. Biofactors 2005; 25(1-4): 87-95.
- Jiang L, Yin X, Chen YH, et al. Proteomic analysis reveals ginsenoside Rb1 attenuates myocardial ischemia/reperfusion injury through inhibiting ROS production from mitochondrial complex I. Theranostics 2021; 11(4): 1703-1720.
- Jardim KV, Siqueira JLN, Báo SN, et al. The role of the lecithin addition in the properties and cytotoxic activity of chitosan and chondroitin sulfate nanoparticles containing curcumin. Carbohydrate Polymers 2020; 227: 115351.
- Bot F, Cossuta D, O'Mahony JA. Inter-relationships between composition, physicochemical properties and functionality of lecithin ingredients. Trends in Food Science & Technology 2021; 111: 261-270.
- Rego RL, Foster NR, Smyrk TC, et al. Prognostic effect of activated EGFR expression in human colon carcinomas: comparison with EGFR status. British Journal of Cancer 2010; 102(1): 165-172.
- Nwaokorie A, Fey D. Personalised Medicine for Colorectal Cancer Using Mechanism-Based Machine Learning Models. International Journal of Molecular Sciences 2021; 22(18): 9970.
- Morteau O, Morham SG, Sellon R, et al. Impaired mucosal defense to acute colonic injury in mice lacking cyclooxygenase-1 or cyclooxygenase-2. The Journal of Clinical Investigation 2000; 105(4): 469-478.
- Zahedi T, Colagar AH, Mahmoodzadeh H. PTGS2 Over-Expression: A Colorectal Carcinoma Initiator not an Invasive Factor. Reports of Biochemistry & Molecular Biology 2021; 9(4): 442-451.
- Ashrafizadeh M, Zarrabi A, Hushmandi K, et al. MicroRNAs in cancer therapy: Their involvement in oxaliplatin sensitivity/resistance of cancer cells with a focus on colorectal cancer. Life Sciences 2020; 256: 117973.
- Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009; 58(10): 1375-1381.
- Li H, Liang J, Wang J, et al. Mex3a promotes oncogenesis through the RAP1/MAPK signaling pathway in colorectal cancer and is inhibited by hsa-miR-6887-3p. Cancer Communications 2021; 41(6): 472-491.
- Luck K, Kim DK, Lambourne L, et al. A reference map of the human binary protein interactome. Nature 2020; 580(7803): 402-408.
- Graves-Deal R, Bogatcheva G, Rehman S, et al. Broad-spectrum receptor tyrosine kinase inhibitors overcome de novo and acquired modes of resistance to EGFR-targeted therapies in colorectal cancer. Oncotarget 2019; 10(13): 1320-1333.
- Zhou J, Ji Q, Li Q. Resistance to anti-EGFR therapies in metastatic colorectal cancer: underlying mechanisms and reversal strategies. Journal of Experimental & Clinical Cancer Research 2021; 40(1): 328.